Pathogen adaptation to vaccination: the Australian Bordetella pertussis story
Laurence Don Wai LuuSchool of Biotechnology and Biomolecular Sciences
University of New South Wales
Sydney, NSW, Australia
Tel: +61 2 9385 3063
Fax: +61 2 9385 1483
Email: laurence.luu@unsw.edu.au
Microbiology Australia 40(4) 177-180 https://doi.org/10.1071/MA19052
Published: 8 November 2019
Whooping cough (pertussis) is a highly contagious vaccine preventable respiratory disease caused by the Gram-negative bacterium Bordetella pertussis. Despite high level vaccination coverage over the past 20 years, Australia has one of the highest per capita burdens of pertussis globally. One of the primary factors associated with the re-emergence of pertussis is pathogen adaptation of B. pertussis to the current acellular vaccines used. This article will focus on the genomic and proteomic changes that have occurred in the Australian B. pertussis population, the significance of these adaptive changes on fitness in a vaccinated environment and what we can do to reduce the significant burden of pertussis in the future.
The rising incidence of B. pertussis in Australia
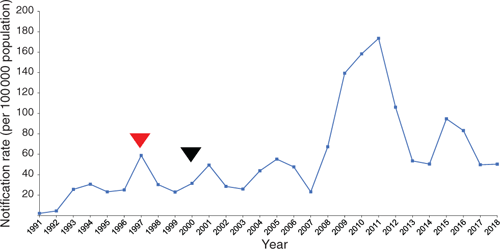
Pertussis vaccinations were first introduced in Australia in 1953 using a whole cell vaccine (WCV), which contained dead B. pertussis cells. This led to a dramatic reduction in the number of pertussis notifications from 767 cases per 100 000 in the 1930s to one case per 100 000 by the late 1970s1. Despite the success of WCVs in reducing pertussis, reports of possible severe side-effects reduced public confidence in the vaccine and led to the replacement of the WCV with a new acellular vaccine (ACV)2.
ACV was introduced into Australia in 1997, initially as a booster, and by 2000 it was used for all immunisations2. There are two ACVs used in Australia, the three component-ACV and the less widely used five-component ACV. The three-component ACV targets three virulence factors; pertussis toxin (Ptx), pertactin (Prn) and filamentous haemagglutinin (Fha), while the five-component vaccine targets two additional fimbriae (Fim2 and Fim3). Since the late 1990s, there has been a steady increase in pertussis notifications observed in Australia and in 2008–2012, Australia experienced a prolonged pertussis epidemic (Figure 1). At the height of the epidemic in 2011, 174 cases per 100 000 were recorded, the highest number documented since the introduction of pertussis vaccination2. This was followed by a smaller yet still significant epidemic from 2014-2017 with a peak of 95 cases per 100 000 in 20153 (Figure 1). Similar trends in increased notification rates have been noted in other countries that switched to the ACV4–6.
|
The changing population structure of Australian B. pertussis in different vaccine eras
To determine how B. pertussis is adapting to vaccine selection pressure, molecular epidemiology typing studies were performed to define the population structure and trace the evolution of B. pertussis in Australia from the pre-vaccination and WCV era to the ACV era.
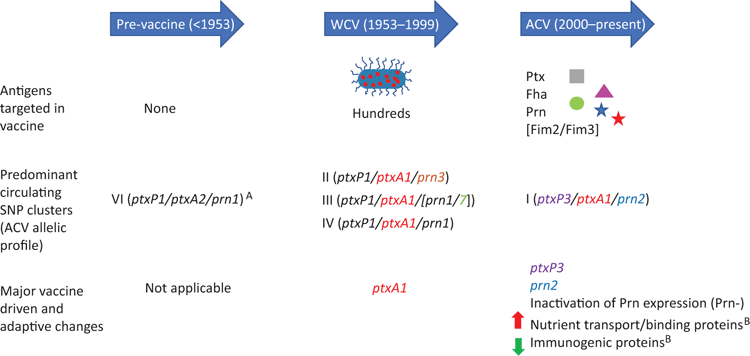
A previous single nucleotide polymorphism (SNP) typing study separated the global pertussis population into 6 SNP clusters (I-VI) based on 65 SNPs7. Australian B. pertussis isolates were mostly found in SNP clusters I-IV with SNP cluster V being a minor Australian cluster and SNP cluster VI containing vaccine and pre-vaccine strains. In the WCV period, SNP cluster II was the predominant cluster and was comprised of 33% of strains typed. After the switch to the ACV, the frequency of SNP cluster II isolates decreased to 11% and was replaced with SNP cluster I, which increased to 31% after it emerged during the WCV/ACV transition period7 (Figure 2). Since the 2008–2012 epidemic, the majority (>90%) of circulating strains in Australia belong to SNP cluster I3,8. Most current circulating strains in other ACV countries typed as ptxP3 strains are equivalent to SNP cluster I9.
Genetic changes in ACV antigen genes of the circulating Australian B. pertussis population
The replacement of the WCV with ACV in Australia reduced the number of antigens targeted from hundreds to 3-5 (Figure 2). This has placed greater selection pressure on genes encoding ACV antigens to change and allow for vaccine escape variants to emerge in Australia. There are two major evolutionary changes observed due to increased vaccine selection pressure: ACV allelic divergence and inactivation of ACV antigen expression (Figure 2).
The greatest ACV allelic divergence is seen in ptxA and prn alleles. SNP cluster I carries the prn2 and ptxA1 alleles which encode antigenic variants that differ from the prn1 and ptxA2 alleles in Tohama I, the strain used to produce the WCV and ACV. The ptxA1 allele emerged prior to the introduction of WCV but only expanded after WCVs were used4 and is present in all cluster I-IV strains. Similarly the prn2 allele emerged during the WCV period in SNP cluster I but expanded after the introduction of ACV4. These mutations in ptxA1 and prn2 alter amino acid residues in known B/T immune epitopes thereby altering recognition from ACV-generated immunity4,10. A double allelic exchange mutant of ACV alleles prn1 and ptxA2 with non-ACV alleles prn2 and ptxA1 conferred a greater survival rate in ACV immunised mice than the wild type strain demonstrating the selective advantage of antigen mismatch in a highly vaccinated environment11.
Besides mismatches in ACV antigens, the current predominance of SNP cluster I is also associated with carrying the ptxP3 allele which encodes a variant pertussis toxin promoter12. This new promoter is associated with increased pertussis toxin production, and possibly virulence and disease severity compared with strains carrying the original ptxP1 allele13,14. Increased pertussis toxin production is thought to provide a selective advantage by delaying neutrophil recruitment and modulating the immune response15. Using a mixed-infection mouse model, SNP cluster I strains outcompeted SNP cluster II strains (carrying ptxP1, prn3 and ptxA1) in a vaccinated environment16. This demonstrates that these genomic changes have increased the overall fitness of the circulating population of B. pertussis and contribute to the predominance of SNP cluster I in an ACV environment.
Recently, strains which do not express ACV antigens have emerged in many developed countries, most notably the Prn deficient strains5,6,17. In Australia, Prn deficient strains are primarily found in SNP cluster I and were first detected at the start of the 2008 epidemic, making up 5% of strains isolated18. However, within a decade this increased to 90% by 2017, the highest proportion of Prn deficient strains in the world3. Over 20 independent mechanisms for Prn inactivation have been documented5,6,18. This diversity of inactivation suggests a beneficial selection pressure for convergent evolution and that Prn deficient strains emerged independently from multiple different clones19. Findings from multiple mouse studies support increased fitness for losing pertactin in an ACV environment with higher survival of Prn deficient strains compared to Prn producing strains in ACV-vaccinated mouse20,21. Prn deficient strains also displayed no differences in disease severity compared to Prn producing strains but have a higher likelihood to cause disease and persist longer in ACV vaccinated individuals14,22. Furthermore, in a mixed infection mouse model, Prn deficient strains had poorer survival in unvaccinated mice compared to Prn producing strains therefore providing evidence that Prn inactivation is an ACV-driven phenomenon. Finally, inactivation of Fha has also been detected in Australia3 while Ptx inactivation has been reported in other countries17. However, these mutant strains are rare, and it remains to be seen whether the loss of these virulence factors increases the fitness of B. pertussis in an ACV environment.
Proteomic changes and broadening the view of pathogen adaptation to vaccination
In addition to increased fitness in an ACV environment, it was shown that SNP cluster I also outcompetes SNP cluster II in an unvaccinated environment16. This suggests that there are other changes between the two clusters that contribute to the current predominance of SNP cluster I in Australia. In proteomic studies, our laboratory identified increased expression of previously unknown adaptations in SNP cluster I of reduced expression of immunogenic proteins such as the type III secretion system and upregulation of amino acid and metal ion transport proteins and adhesins23,24 (Figure 2). Some of these proteomic differences were associated with genetic mutations which are also found in other global pertussis strains23. Additionally, several other transcriptomic studies have reported gene expression differences associated with the current epidemic pertussis strains25,26. Together, these studies have broadened our understanding of how B. pertussis is evolving and identified additional pathogen factors important for the re-emergence of pertussis.
Future strategies to reduce the burden of pertussis
The ongoing evolution of B. pertussis in response to vaccine selection requires continued long-term epidemiological surveillance in Australia to monitor vaccine escape strains and the possible introduction of emerging antibiotic resistant B. pertussis strains from other countries27. Although B. pertussis is adapting to the ACV, the current ACV remains effective in preventing pertussis disease in fully immunised individuals28. Additionally, maternal immunisation and cocooning are also effective strategies at protecting the most vulnerable newborns from pertussis29,30. However, for long term prevention and protection, an improved vaccine is required in the future. Current proposed strategies to alter vaccine alleles to better match SNP cluster I will improve our ability to target these strains but will not be sufficient to combat Prn deficient strains. Therefore, a future pertussis vaccine should broaden the number of antigens targeted and ensure that these antigens are essential to B. pertussis survival so as to limit further pathogen adaptation. To do this, further research is required to better understand fundamental aspects of pertussis biology and adaptation as well as the processes behind vaccine and host-induced immunity.
Conflicts of interest
The author declares no conflicts of interest.
Acknowledgements
Laurence Don Wai Luu was supported by an Australian Government Research Training Program Scholarship and an NHMRC project grant awarded to Professor Ruiting Lan. The author thanks Dr Sophie Octavia and Professor Ruiting Lan for providing feedback on the manuscript.
References
[1] Hall, R. (1993) Notifiable diseases surveillance, 1917 to 1991. Commun. Dis. Intell. 17, 226–236.[2] Pillsbury, A. et al. (2014) Australian vaccine preventable disease epidemiological review series: pertussis. Commun. Dis. Intell. 38, E179–E194.
[3] Xu, Z. et al. (2019) Pertactin-negative and filamentous hemagglutinin-negative Bordetella pertussis, Australia, 2013–2017. Emerg. Infect. Dis. 25, 1196.
| Pertactin-negative and filamentous hemagglutinin-negative Bordetella pertussis, Australia, 2013–2017.Crossref | GoogleScholarGoogle Scholar | 31107218PubMed |
[4] Bart, M.J. et al. (2014) Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio 5, e01074-14.
| Global population structure and evolution of Bordetella pertussis and their relationship with vaccination.Crossref | GoogleScholarGoogle Scholar | 24757216PubMed |
[5] Otsuka, N. et al. (2012) Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One 7, e31985.
| Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan.Crossref | GoogleScholarGoogle Scholar | 22348138PubMed |
[6] Barkoff, A.-M. et al. (2019) Pertactin-deficient Bordetella pertussis isolates: evidence of increased circulation in Europe, 1998 to 2015. Euro Surveill. 24, pii=1700832.
| Pertactin-deficient Bordetella pertussis isolates: evidence of increased circulation in Europe, 1998 to 2015.Crossref | GoogleScholarGoogle Scholar | 30782265PubMed |
[7] Octavia, S. et al. (2011) Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection. Mol. Biol. Evol. 28, 707–715.
| Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection.Crossref | GoogleScholarGoogle Scholar | 20833694PubMed |
[8] Octavia, S. et al. (2012) Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010. J. Infect. Dis. 205, 1220–1224.
| Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010.Crossref | GoogleScholarGoogle Scholar | 22416243PubMed |
[9] van Gent, M. et al. (2012) Small mutations in Bordetella pertussis are associated with selective sweeps. PLoS One 7, e46407.
| Small mutations in Bordetella pertussis are associated with selective sweeps.Crossref | GoogleScholarGoogle Scholar | 23029513PubMed |
[10] He, Q. et al. (2003) Bordetella pertussis protein pertactin induces type-specific antibodies: one possible explanation for the emergence of antigenic variants? J. Infect. Dis. 187, 1200–1205.
| Bordetella pertussis protein pertactin induces type-specific antibodies: one possible explanation for the emergence of antigenic variants?Crossref | GoogleScholarGoogle Scholar | 12695998PubMed |
[11] Komatsu, E. et al. (2010) Synergic effect of genotype changes in pertussis toxin and pertactin on adaptation to an acellular pertussis vaccine in the murine intranasal challenge model. Clin. Vaccine Immunol. 17, 807–812.
| Synergic effect of genotype changes in pertussis toxin and pertactin on adaptation to an acellular pertussis vaccine in the murine intranasal challenge model.Crossref | GoogleScholarGoogle Scholar | 20357056PubMed |
[12] Lam, C. et al. (2012) Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis. Infect. Genet. Evol. 12, 492–495.
| Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis.Crossref | GoogleScholarGoogle Scholar | 22293463PubMed |
[13] Mooi, F.R. et al. (2009) Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg. Infect. Dis. 15, 1206.
| Bordetella pertussis strains with increased toxin production associated with pertussis resurgence.Crossref | GoogleScholarGoogle Scholar | 19751581PubMed |
[14] Clarke, M. et al. (2016) The relationship between Bordetella pertussis genotype and clinical severity in Australian children with pertussis. J. Infect. 72, 171–178.
| The relationship between Bordetella pertussis genotype and clinical severity in Australian children with pertussis.Crossref | GoogleScholarGoogle Scholar | 26675318PubMed |
[15] Kirimanjeswara, G.S. et al. (2005) Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis. J. Clin. Invest. 115, 3594.
| Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis.Crossref | GoogleScholarGoogle Scholar | 16294220PubMed |
[16] Safarchi, A. et al. (2016) Better colonisation of newly emerged Bordetella pertussis in the co-infection mouse model study. Vaccine 34, 3967–3971.
| Better colonisation of newly emerged Bordetella pertussis in the co-infection mouse model study.Crossref | GoogleScholarGoogle Scholar | 27346304PubMed |
[17] Bouchez, V. et al. (2009) First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin. Vaccine 27, 6034–6041.
| First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin.Crossref | GoogleScholarGoogle Scholar | 19666155PubMed |
[18] Lam, C. et al. (2014) Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg. Infect. Dis. 20, 626.
| Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia.Crossref | GoogleScholarGoogle Scholar | 24655754PubMed |
[19] Safarchi, A. et al. (2016) Genomic dissection of Australian Bordetella pertussis isolates from the 2008–2012 epidemic. J. Infect. 72, 468–477.
| Genomic dissection of Australian Bordetella pertussis isolates from the 2008–2012 epidemic.Crossref | GoogleScholarGoogle Scholar | 26826518PubMed |
[20] Safarchi, A. et al. (2015) Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model. Vaccine 33, 6277–6281.
| Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model.Crossref | GoogleScholarGoogle Scholar | 26432908PubMed |
[21] Hegerle, N. et al. (2014) Pertactin deficient Bordetella pertussis present a better fitness in mice immunized with an acellular pertussis vaccine. Vaccine 32, 6597–6600.
| Pertactin deficient Bordetella pertussis present a better fitness in mice immunized with an acellular pertussis vaccine.Crossref | GoogleScholarGoogle Scholar | 25312274PubMed |
[22] Martin, S.W. et al. (2015) Pertactin-negative Bordetella pertussis strains: evidence for a possible selective advantage. Clin. Infect. Dis. 60, 223–227.
| Pertactin-negative Bordetella pertussis strains: evidence for a possible selective advantage.Crossref | GoogleScholarGoogle Scholar | 25301209PubMed |
[23] Luu, L.D.W. et al. (2018) Comparison of the whole cell proteome and secretome of epidemic Bordetella pertussis strains from the 2008–2012 Australian epidemic under sulfate modulating conditions. Front. Microbiol. 9, 2851.
| Comparison of the whole cell proteome and secretome of epidemic Bordetella pertussis strains from the 2008–2012 Australian epidemic under sulfate modulating conditions.Crossref | GoogleScholarGoogle Scholar |
[24] Luu, L.D.W. et al. (2018) Proteomic adaptation of Australian epidemic Bordetella pertussis. Proteomics 18, e1700237.
| Proteomic adaptation of Australian epidemic Bordetella pertussis.Crossref | GoogleScholarGoogle Scholar |
[25] King, A.J. et al. (2013) Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains. PLoS One 8, e66150.
| Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains.Crossref | GoogleScholarGoogle Scholar | 24324664PubMed |
[26] de Gouw, D. et al. (2014) Differentially expressed genes in Bordetella pertussis strains belonging to a lineage which recently spread globally. PLoS One 9, e84523.
| Differentially expressed genes in Bordetella pertussis strains belonging to a lineage which recently spread globally.Crossref | GoogleScholarGoogle Scholar | 25133400PubMed |
[27] Xu, Z. et al. (2019) Genomic epidemiology of erythromycin-resistant Bordetella pertussis in China. Emerg. Microbes Infect. 8, 461–470.
| Genomic epidemiology of erythromycin-resistant Bordetella pertussis in China.Crossref | GoogleScholarGoogle Scholar | 30898080PubMed |
[28] Warfel, J.M. et al. (2014) Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc. Natl. Acad. Sci. USA 111, 787–792.
| Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model.Crossref | GoogleScholarGoogle Scholar | 24277828PubMed |
[29] Amirthalingam, G. et al. (2014) Effectiveness of maternal pertussis vaccination in England: an observational study. Lancet 384, 1521–1528.
| Effectiveness of maternal pertussis vaccination in England: an observational study.Crossref | GoogleScholarGoogle Scholar | 25037990PubMed |
[30] Rowe, S.L. et al. (2018) Effectiveness of parental cocooning as a vaccination strategy to prevent pertussis infection in infants: a case-control study. Vaccine 36, 2012–2019.
| Effectiveness of parental cocooning as a vaccination strategy to prevent pertussis infection in infants: a case-control study.Crossref | GoogleScholarGoogle Scholar | 29525284PubMed |
Biography
Laurence Don Wai Luu is a postdoctoral research associate in Professor Ruiting Lan’s lab at the University of New South Wales. His research interest involves the application of genomics and proteomics to better understand the biology and evolution of Bordetella pertussis.


 ) and exclusive use of ACV for all vaccinations after 2000 (▾) are indicated. Data from the Department of Health, 2019.
) and exclusive use of ACV for all vaccinations after 2000 (▾) are indicated. Data from the Department of Health, 2019.