Corynebacterium and Dolosigranulum: future probiotic candidates for upper respiratory tract infections
Rachael Lappan A B D and Christopher S Peacock A CA The Marshall Centre for Infectious Diseases Research and Training, School of Biomedical Sciences, The University of Western Australia, Perth, WA, Australia
B Wesfarmers Centre of Vaccines and Infectious Diseases, Telethon Kids Institute, The University of Western Australia, Perth, WA, Australia
C Telethon Kids Institute, The University of Western Australia, Perth, WA, Australia. Tel: +61 8 6457 6172, Email: Christopher.Peacock@uwa.edu.au
D Present address: The School of Biological Sciences, 18 Innovation Walk, Monash University, Clayton, Vic. 3800, Australia. Tel: +61 3 9905 5794, Email: Rachael.Lappan@monash.edu
Microbiology Australia 40(4) 172-177 https://doi.org/10.1071/MA19051
Published: 11 November 2019
The presence of the bacterial genera Corynebacterium and Dolosigranulum has consistently been associated with a healthy upper respiratory tract (URT). Commonly occurring together in the nasopharynx of healthy children, the role of these commensal organisms in nasopharyngeal health is unknown, as few studies have sought to determine whether they actively contribute to maintaining a healthy state. We recently identified Corynebacterium pseudodiphtheriticum and Dolosigranulum pigrum as the major nasopharyngeal species associated with resistance to recurrent ear infections, via 16S rRNA gene sequencing and metagenomics. Using in vitro bacterial interference assays, we observed a reduction in the growth of Moraxella catarrhalis – one of the three major otopathogens – in the presence of C. pseudodiphtheriticum. Further in vitro and in vivo studies of the interactions between commensal C. pseudodiphtheriticum and D. pigrum strains, URT pathogens, and the human host will help to clarify their role in nasopharyngeal health. If they play a protective role, these organisms are promising candidates for the development of a probiotic therapy for the treatment or prevention of URT diseases in children.
Human microbiome research has traditionally focused on differences in the composition of the microbial community between healthy and disease states. This research has naturally progressed into ‘mining the microbiota for therapeutics’; the process of identifying members of the microbiota that contribute to health and developing them into therapeutic agents, either in the form of probiotics (live, beneficial microorganisms) or isolated antimicrobial compounds. Several examples mirroring this process have already been demonstrated, where defined bacterial mixtures have successfully treated1 or prevented2 infection, or the production of an antimicrobial substance by a commensal organism inhibits a pathogenic close relative3,4.
Markers of a healthy nasopharynx
In the microbiome of the upper respiratory tract (URT), the genera Corynebacterium and Dolosigranulum have been consistently associated with health, particularly in children (Table 1). In these studies, they are frequently reported to co-occur, their presence corresponds with breastfeeding and a lower risk of respiratory infection, and they appear to be negatively impacted by antibiotic use. This suggests that these genera are important characteristics of a healthy nasopharynx. It may be hypothesised that the absence or reduction (by antibiotic use) of Corynebacterium and Dolosigranulum from the URT microbiota results in respiratory pathogens preferentially colonising the nasopharynx and an increased susceptibility to respiratory disease.
|
While it is clear that Corynebacterium and Dolosigranulum are strongly associated with health, little work has been done in vitro to characterise how they contribute to health. Do they compete with respiratory pathogens for nutrients and space, or prevent infection via bacterial antagonism? No previous studies appear to have directly tested D. pigrum for such activity. Of those that have investigated Corynebacterium, one early study reported C. pseudodiphtheriticum binding to pharyngeal cells, competitively excluding M. catarrhalis; though no zone of inhibition was observed on blood agar18. A more recent study reported that inoculating C. pseudodiphtheriticum strain 090104 into mice improved resistance to infection by respiratory syncytial virus and secondary pneumococcal infection, where non-viable 090104 had a weaker effect19. Recently, the commensal also demonstrated inhibition of S. aureus by exploiting its own virulence components20. Other Corynebacterium species have shown similar promising antimicrobial activity; C. propinquum uses siderophores to restrict the availability of iron to coagulase-negative staphylococci21, and C. accolens inhibits S. pneumoniae via antimicrobial free fatty acids, which C. accolens requires for growth10.
C. pseudodiphtheriticum and D. pigrum in resistance to recurrent ear infections
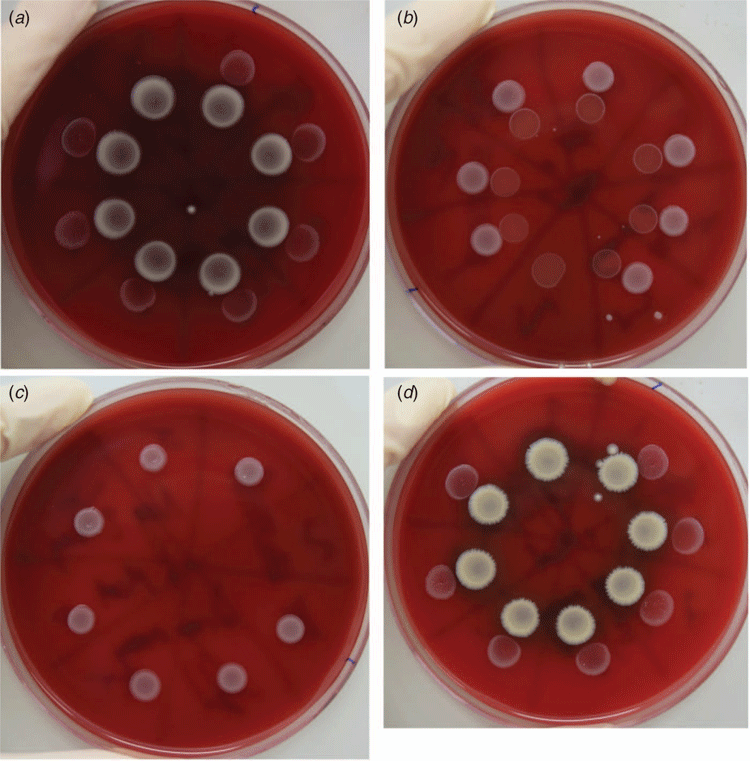
Recently, we used 16S rRNA gene sequencing to characterise the nasopharyngeal microbiomes of children with recurrent ear infections (recurrent acute otitis media; rAOM), and healthy rAOM-resistant children17. The relative abundance of Corynebacterium and Dolosigranulum was significantly higher in the rAOM-resistant children, concordant with previous studies comparing the nasopharyngeal microbiota of children with and without OM6,7,13. Metagenomics further revealed that C. pseudodiphtheriticum and D. pigrum were the dominant species in the healthy rAOM-resistant nasopharynx with C. propinquum and C. accolens present to a lesser extent22. To investigate whether these commensal organisms have a protective role in the nasopharynx interfering with the growth of pathogens, we conducted bacterial interference assays22. C. pseudodiphtheriticum and D. pigrum were assessed for inhibitory activity against the major otopathogen species (non-typeable Haemophilus influenzae – NTHi, Streptococcus pneumoniae and Moraxella catarrhalis); two strains of each commensal species and seven strains of each otopathogen were tested, including mixtures of both commensals together. Growing the pathogens adjacent to the commensals on agar revealed that neither commensal affected the growth of S. pneumoniae or NTHi, but all seven M. catarrhalis strains were inhibited by C. pseudodiphtheriticum (Figure 1). This effect was not influenced by the presence of D. pigrum.
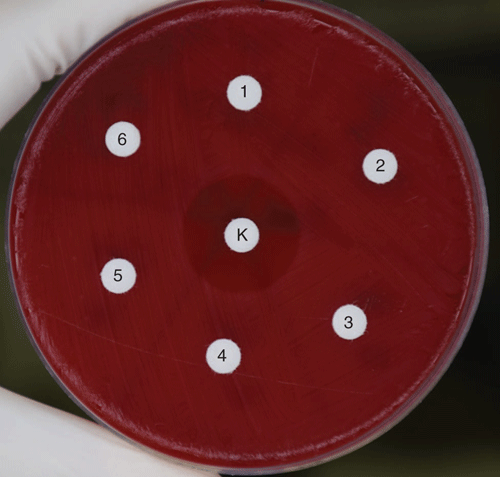
To attempt to identify the production of an antimicrobial compound by C. pseudodiphtheriticum as a potential mechanism, we inoculated filter paper discs with cell-free C. pseudodiphtheriticum broth culture and cell lysates for a disc diffusion assay on M. catarrhalis. No inhibition was observed (Figure 2)22. It is possible that the inhibitory effect of C. pseudodiphtheriticum is not mediated by an antimicrobial substance, for example by competitive acquisition of nutrients. However, it is also likely that such a substance is produced, but was not successfully extracted from C. pseudodiphtheriticum culture in a functional state or at sufficient concentrations to produce a zone of inhibition, or its production requires a trigger (like the presence of M. catarrhalis). This preliminary work demonstrates that C. pseudodiphtheriticum inhibits the growth of M. catarrhalis; while this is the least common of the major otopathogens, it may be an important one to target given its apparent synergistic interactions with NTHi and S. pneumoniae23,24. However, the mechanism of interference, the role of D. pigrum and whether Corynebacterium species can inhibit URT pathogens at a clinically useful degree is yet unknown.
|
Towards a potential probiotic therapy using Corynebacterium and Dolosigranulum
The consistent correlation between Corynebacterium, Dolosigranulum and upper respiratory health presents a very promising avenue of research towards development of probiotic therapies for the URT. The recent studies demonstrating antimicrobial activity by C. accolens against S. pneumoniae10, C. pseudodiphtheriticum against both S. aureus20 and M. catarrhalis22, and C. propinquum against coagulase-negative staphylococci21 suggest it is possible that multiple Corynebacterium species are beneficial to the URT, acting via different mechanisms against different pathogens. The role of D. pigrum in these interactions remains unknown, however it has been hypothesised that the production of lactic acid by D. pigrum lowers the pH of the local environment, selecting for the growth of Corynebacterium species25. This is highly plausible, as several species of Lactobacillus inhibit M. catarrhalis via lactic acid26, which also exhibits antimicrobial and anti-inflammatory properties in the human vagina27. A niche-specific probiotic therapy containing multiple species with complementary effects may be an effective approach for the URT, especially for restoration after antibiotic use, which has been demonstrated to reduce Corynebacterium and Dolosigranulum populations (Table 1). Previous clinical trials using nasal spray probiotics for children with rAOM have suggested that pre-treatment with antibiotics allows re-establishment of the healthy microbiota, preventing the re-emergence of pathogens28,29.
To fully investigate the possibility that these commensal organisms work synergistically to provide resistance to respiratory pathogens, and the potential for this to be translated into a therapy, there is an array of possibilities for future research. First, it remains likely that C. pseudodiphtheriticum produces an antimicrobial substance that inhibits M. catarrhalis. To isolate it, a range of commensal nasopharyngeal strains could be screened for optimal antimicrobial activity, and optimisation of culture conditions or enrichment of proteins may be required, similar to the approach taken by Latham et al. (2017) for the isolation of an antimicrobial substance from Haemophilus haemolyticus4. The commensal Corynebacterium genomes are not well characterised; a very recent sequencing effort by Stubbendieck et al. (2019) has improved the available genomic information for C. pseudodiphtheriticum and C. propinquum21, but these draft genome assemblies are still incomplete as of August 2019 (NCBI Genome Assembly and Annotation reports). There is now one strain of D. pigrum with a complete assembly (83VPs-KB5) available. Mining the genomes of multiple commensal strains for putative antimicrobial genes may provide further evidence for protective factors. Additionally, epithelial cell models have previously been successful in understanding the competition between H. haemolyticus and NTHi in the URT30 and would also be useful to obtain a deeper insight into the competitive mechanisms and host responses, to see if colonisation with these organisms influences susceptibility to later infection with S. pneumoniae, M. catarrhalis or S. aureus.
Further research into the potential of Corynebacterium and Dolosigranulum to compete with respiratory pathogens, alone or in synergy, will provide a strong direction for the use of animal models and clinical trials in the development of a probiotic therapy, which has the potential to treat or prevent a range of upper respiratory illnesses in children.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Kyra Cottrell, Hanna Sidjabat, Janessa Pickering and Barbara Chang for helpful discussion on the bacterial interference assays. We also thank Lea-Ann Kirkham, Caitlyn Granland, Deborah Lehmann and Jacinta Bowman for the provision of the otopathogen strains, and Paul Healy for the provision of the C. pseudodiphtheriticum and D. pigrum strains. This research was supported by the Telethon New Childrens’ Hospital Research Fund.
References
[1] Lawley, T.D. et al. (2012) Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995.| Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice.Crossref | GoogleScholarGoogle Scholar | 23133377PubMed |
[2] Brugiroux, S. et al. (2016) Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 16215.
| Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium.Crossref | GoogleScholarGoogle Scholar | 27869789PubMed |
[3] Iwase, T. et al. (2010) Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349.
| Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization.Crossref | GoogleScholarGoogle Scholar | 20485435PubMed |
[4] Latham, R.D. et al. (2017) An isolate of Haemophilus haemolyticus produces a bacteriocin-like substance that inhibits the growth of nontypeable Haemophilus influenzae. Int. J. Antimicrob. Agents 49, 503–506.
| An isolate of Haemophilus haemolyticus produces a bacteriocin-like substance that inhibits the growth of nontypeable Haemophilus influenzae.Crossref | GoogleScholarGoogle Scholar | 28242259PubMed |
[5] Bogaert, D. et al. (2011) Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6, e17035.
| Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis.Crossref | GoogleScholarGoogle Scholar | 21695210PubMed |
[6] Laufer, A.S. et al. (2011) Microbial communities of the upper respiratory tract and otitis media in children. MBio 2, e00245-10.
| Microbial communities of the upper respiratory tract and otitis media in children.Crossref | GoogleScholarGoogle Scholar | 21285435PubMed |
[7] Pettigrew, M.M. et al. (2012) Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78, 6262–6270.
| Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children.Crossref | GoogleScholarGoogle Scholar | 22752171PubMed |
[8] Biesbroek, G. et al. (2014) The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 190, 298–308.
| The impact of breastfeeding on nasopharyngeal microbial communities in infants.Crossref | GoogleScholarGoogle Scholar | 24921688PubMed |
[9] Biesbroek, G. et al. (2014) Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 190, 1283–1292.
| Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children.Crossref | GoogleScholarGoogle Scholar | 25329446PubMed |
[10] Bomar, L. et al. (2016) Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio 7, e01725-15.
| Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols.Crossref | GoogleScholarGoogle Scholar | 26733066PubMed |
[11] Prevaes, S.M.P.J. et al. (2016) Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 193, 504–515.
| Development of the nasopharyngeal microbiota in infants with cystic fibrosis.Crossref | GoogleScholarGoogle Scholar |
[12] Bosch, A.A.T.M. et al. (2017) Maturation of the infant respiratory microbiota, environmental drivers, and health consequences: a prospective cohort study. Am. J. Respir. Crit. Care Med. 196, 1582–1590.
| Maturation of the infant respiratory microbiota, environmental drivers, and health consequences: a prospective cohort study.Crossref | GoogleScholarGoogle Scholar |
[13] Chonmaitree, T. et al. (2017) Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One 12, e0180630.
| Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media.Crossref | GoogleScholarGoogle Scholar | 28708872PubMed |
[14] Hasegawa, K. et al. (2017) Nasal airway microbiota profile and severe bronchiolitis in infants: a case-control study. Pediatr. Infect. Dis. J. 36, 1044–1051.
| Nasal airway microbiota profile and severe bronchiolitis in infants: a case-control study.Crossref | GoogleScholarGoogle Scholar | 28005692PubMed |
[15] Kelly, M.S. et al. (2017) The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr. Infect. Dis. J. 36, e211–e218.
| The nasopharyngeal microbiota of children with respiratory infections in Botswana.Crossref | GoogleScholarGoogle Scholar | 28399056PubMed |
[16] Copeland, E. et al. (2018) Chronic rhinosinusitis: potential role of microbial dysbiosis and recommendations for sampling sites. Front. Cell. Infect. Microbiol. 8, 57.
| Chronic rhinosinusitis: potential role of microbial dysbiosis and recommendations for sampling sites.Crossref | GoogleScholarGoogle Scholar | 29541629PubMed |
[17] Lappan, R. et al. (2018) A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 18, 13.
| A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera.Crossref | GoogleScholarGoogle Scholar | 29458340PubMed |
[18] Rikitomi, N. et al. (1989) Role of normal microflora in the throat in inhibition of adherence of pathogenic bacteria to host cells: in vitro competitive adherence between Corynebacterium pseudodiphtheriticum and Branhamella catarrhalis. Kansenshogaku Zasshi 63, 118–124.
| Role of normal microflora in the throat in inhibition of adherence of pathogenic bacteria to host cells: in vitro competitive adherence between Corynebacterium pseudodiphtheriticum and Branhamella catarrhalis.Crossref | GoogleScholarGoogle Scholar | 2501428PubMed |
[19] Kanmani, P. et al. (2017) Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae Superinfection. Front. Microbiol. 8, 1613.
| Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae Superinfection.Crossref | GoogleScholarGoogle Scholar | 28878760PubMed |
[20] Hardy, B.L. et al. (2019) Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. MBio 10, e02491-18.
| Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy.Crossref | GoogleScholarGoogle Scholar | 30622190PubMed |
[21] Stubbendieck, R.M. et al. (2019) Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl. Environ. Microbiol. 85, e02406–e02418.
| 30578265PubMed |
[22] Lappan, R. (2019) Using ’omics technologies to understand pathogenesis and seek alternative therapies for otitis media in children. The University of Western Australia.
[23] Armbruster, C.E. et al. (2010) Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio 1, e00102–e00110.
| Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling.Crossref | GoogleScholarGoogle Scholar | 20802829PubMed |
[24] Perez, A.C. et al. (2014) Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog. Dis. 70, 280–288.
| Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo.Crossref | GoogleScholarGoogle Scholar | 24391058PubMed |
[25] de Steenhuijsen Piters, W.A.A. et al. (2015) The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140294.
| The role of the local microbial ecosystem in respiratory health and disease.Crossref | GoogleScholarGoogle Scholar |
[26] van den Broek, M.F. et al. (2018) Multifactorial inhibition of lactobacilli against the respiratory tract pathogen Moraxella catarrhalis. Benef. Microbes 9, 429–439.
| Multifactorial inhibition of lactobacilli against the respiratory tract pathogen Moraxella catarrhalis.Crossref | GoogleScholarGoogle Scholar | 29633637PubMed |
[27] Tachedjian, G. et al. (2017) The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 168, 782–792.
| The role of lactic acid production by probiotic Lactobacillus species in vaginal health.Crossref | GoogleScholarGoogle Scholar | 28435139PubMed |
[28] Roos, K. et al. (2001) Effect of recolonisation with “interfering” α streptococci on recurrences of acute and secretory otitis media in children: randomised placebo controlled trial. BMJ 322, 210–212.
| Effect of recolonisation with “interfering” α streptococci on recurrences of acute and secretory otitis media in children: randomised placebo controlled trial.Crossref | GoogleScholarGoogle Scholar | 11159619PubMed |
[29] Tano, K. et al. (2002) A nasal spray with alpha-haemolytic streptococci as long term prophylaxis against recurrent otitis media. Int. J. Pediatr. Otorhinolaryngol. 62, 17–23.
| A nasal spray with alpha-haemolytic streptococci as long term prophylaxis against recurrent otitis media.Crossref | GoogleScholarGoogle Scholar | 11738689PubMed |
[30] Pickering, J.L. et al. (2016) Haemophilus haemolyticus interaction with host cells is different to nontypeable Haemophilus influenzae and prevents NTHi association with epithelial cells. Front. Cell. Infect. Microbiol. 6, 50.
| Haemophilus haemolyticus interaction with host cells is different to nontypeable Haemophilus influenzae and prevents NTHi association with epithelial cells.Crossref | GoogleScholarGoogle Scholar | 27242968PubMed |
Biographies
Rachael Lappan completed her PhD on the microbiome of rAOM at the University of Western Australia and Telethon Kids Institute in 2019. She now works as a postdoctoral fellow at Monash University, broadening her horizons with research on enteropathogens and microbial communities in more extreme environments like arid deserts and the aerosphere.
Christopher Peacock, BSc Hons Biological Sciences (1985), Fellow of the Institute of Medical Laboratory Sciences (1988), University of London, PhD Human Genetics on ‘the Susceptibility to Visceral Leishmaniasis’ (1998), University of Cambridge. After running a research and service histology laboratory at the London School of Tropical Medicine and Hygiene, he spent one year on a successful HIV project in Abidjan, West Africa followed by two years in the Amazon region of Brazil working on genetic susceptibility to TB, Leprosy and Leishmaniasis. Having completed his PhD in 1998, he undertook a position as a postdoctoral research associate continuing the work on human susceptibility to infectious diseases followed by a role as a senior computational biologist at the Wellcome Trust Sanger Institute leading to publication of the first Leishmania genome as part a Special edition of Science in 2005 and the publication of a comparative Leishmania genome paper in 2007 published in Nature Genetics. In 2007, he moved to Australia to help set up a Division of Genetics and Health in infectious diseases at the Telethon Institute of Child Health Research and in 2009 took up a senior lecturer position at the University of Western Australia. Shortly afterwards he was awarded one of the inaugural ARC Future Fellowships. In addition to neglected tropical diseases, his research interests now encompass metagenomics, and novel parasitic infections in indigenous wildlife.