Cytotoxic factor influencing acquired antimicrobial resistance in Pseudomonas aeruginosa
Dinesh Subedi A , Ajay Kumar Vijay B , Scott A Rice C and Mark Willcox DA School of Optometry and Vision Science, UNSW Sydney, NSW 2052, Australia. Tel: +61 4 1397 3921, Fax: +61 2 9313 6243, Email: d.subedi@unsw.edu.au, subedi.dnes@gmail.com
B School of Optometry and Vision Science, UNSW Sydney, NSW 2052, Australia. Tel: +61 2 9385 4503, Fax: +61 2 9313 6243, Email: v.ajaykumar@unsw.edu.au
C School of Biological Sciences, NTU. Tel: +65 6592 7944, Office: SBS B1N 27, Email: rscott@ntu.edu.sg
D School of Optometry and Vision Science, UNSW Sydney, NSW 2052, Australia. Tel: +61 2 9385 4164, Email: m.willcox@unsw.edu.au
Microbiology Australia 40(4) 161-164 https://doi.org/10.1071/MA19048
Published: 29 October 2019
The Gram-negative opportunistic bacterium Pseudomonas aeruginosa is associated with different types of human infections and because of emerging multidrug-resistant strains, these infections are of major global public health concern. Certain strains possess a unique cytotoxic effector protein ExoU, which contributes to the fitness of this organism in different ecological niches and is associated with acquired antibiotic resistance. This article summarises the current knowledge of the exoU gene in P. aeruginosa, including genetics, distribution in strains from different locations and association with antibiotic resistance. Understanding of this effector protein may have important implications for the understanding of pathogenesis and antimicrobial resistance in P. aeruginosa infections.
The type III secretion system (TTSS), which injects effector proteins into host cells, is an important determinant of virulence in the opportunistic pathogen Pseudomonas aeruginosa. The bacterium encodes four effector proteins that are secreted by the TTSS: ExoY, ExoS, ExoT and ExoU1. Of note, a complete set of genes for these effector proteins may not be present in all isolates. Almost all strains carry both the exoY and exoT genes. However, a strain may possess either exoU or exoS but very rarely both2,3. ExoU has phospholipase activity that rapidly kills cells and hence is associated with severe disease outcomes4.
The exoU gene was first identified in a highly cytotoxic P. aeruginosa strain PA1033. Using in vitro infection studies, Fleiszig et al. demonstrated that PA103 was highly cytotoxic despite the absence of the exoS gene3. It was subsequently found that the strain encodes a 72 kDa protein that was associated with the cytotoxicity and was named ExoU, after the nomenclature of ExoS and ExoT5. ExoU had been previously identified independently and called PepA (Pseudomonas exoprotein A), although ExoU is now the accepted name. Both studies demonstrated that an exoU knockout strain was non-cytotoxic in in vitro and in vivo infection models5,6. Further studies indicated that exoU is a variable trait amongst clinical isolates of P. aeruginosa6. Several lines of evidence from earlier molecular studies have suggested that exoU and its cognate chaperon spcU are located within a region of the chromosome associated with genomic plasticity and the percent G+C of exoU/spcU (58.8) is less than the average percent G+C content (66.6) of P. aeruginosa5,7. These findings implied that exoU may be a recently acquired gene in P. aeruginosa.
In phylogenetic comparisons, exoU+ strains form a separate clade, which contains less strains than the clade composed of exoS+ strains8,9. There is an unequal distribution of exoU+ strains in different clinical and environmental settings, with the exoU+ strains being less abundant than exoS+ strains. In acute infections, exoU+ strains are isolated from 28–42% of samples whilst in chronic infections, such as lung infection of cystic fibrosis patients, the rate of isolation of exoU+ strains is less than 10%2,4,10. Furthermore, exoU+ strains were less prevalent in environmental samples compared to clinical samples. However, within the samples from the environment, exoU+ strains were over-represented in domestic (e.g. sinks, drains, toilets, fountains, hoses)11 and hospital (e.g. sinks and washtubs of intensive care units)12 environments versus natural environments (e.g. soil and plants).
Perhaps the most intriguing difference in the distribution of exoU+ strains was noted in the strains isolated from corneal ulcers13. Strains possessing exoU have been isolated from corneal samples in 33% to 61% of cases8,14–16,. These rates are higher than those commonly observed in other acute infections. The risk of keratitis caused by P. aeruginosa is linked to contact lens wear. ExoU+ strains are more commonly isolated from keratitis associated with contact lens wear14,15. Furthermore, ulcers caused by exoU+ strains result in worse visual outcomes than those caused by strains lacking exoU16. The main reason for this could be that the exoU+ strains tend to be more prevalent in household tap water including sinks and drains, which could contaminate contact lens as many people insert lenses in the bathroom. However, there is lack of literature defining the roles of environmental reservoirs in dictating the prevalence of exoU+ versus exoS+ strains in keratitis. More studies are needed to confirm the disproportionate distribution of exoU+ and exoS+ strains in environmental reservoirs and fitness of different genotypes in various environments.
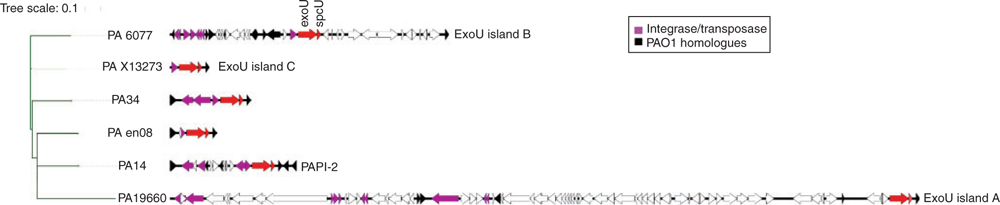
With the availability of metagenome data, the genomic context of the exoU carrying genomic island has been elucidated. The exoU island is derived from the integrative plasmid pKCL102 and is integrated at a tRNA-Lys gene adjacent to the locus PA0976 of PAO117,18. The pKCL102 contains XerC/XerD-integrase, which is responsible for site-specific integration and because of possession of origin of replication (oriV), it can replicate autonomously. Thereby, pKCL102 can occur in multiple copies17. We have previously demonstrated that in the keratitis strain PA34, the exoU gene is found on a 7.5 kb island flanked by tRNA-Lys and homologs of the PAO1 genes PA0976 and PA098819. However, there is a wide variation in the size of the exoU island between strains, for example, it is 14 kb (PAPI-2) in strain PA14 (highly virulent reference strain), 81 kb (exoU island A) in ocular isolate 6077, 29.8 kb (exoU island B) in ocular isolate 19660, 3.9 kb (exoU island C) in blood isolate X13273 and 3.5 kb in an environmental isolate (Figure 1)20,21. Despite the differences in the size of exoU carrying islands between strains, the exoU gene appears to encode a functional cytotoxin19. The difference in island size is attributed to the presence of several mobile elements including insertion sequences and transposons, which can be subjected to recombination, deletion or elimination in response to the environmental selective pressure. It has been hypothesised that such mobile elements potentially lead to excision of exoS leaving the strain as exoU+ (cytotoxic)21. However, exoU and exoS are not closely located on the chromosome and no explanation has emerged for strains that carry either both exoU and exoS or neither. More importantly, understanding the selection pressures such as growth in the presence of predators (e.g. protozoa) or antimicrobials that might favour the acquisition of exoU in different environmental settings will help to better understand pathogenesis and epidemiology of different types of P. aeruginosa infections.
|
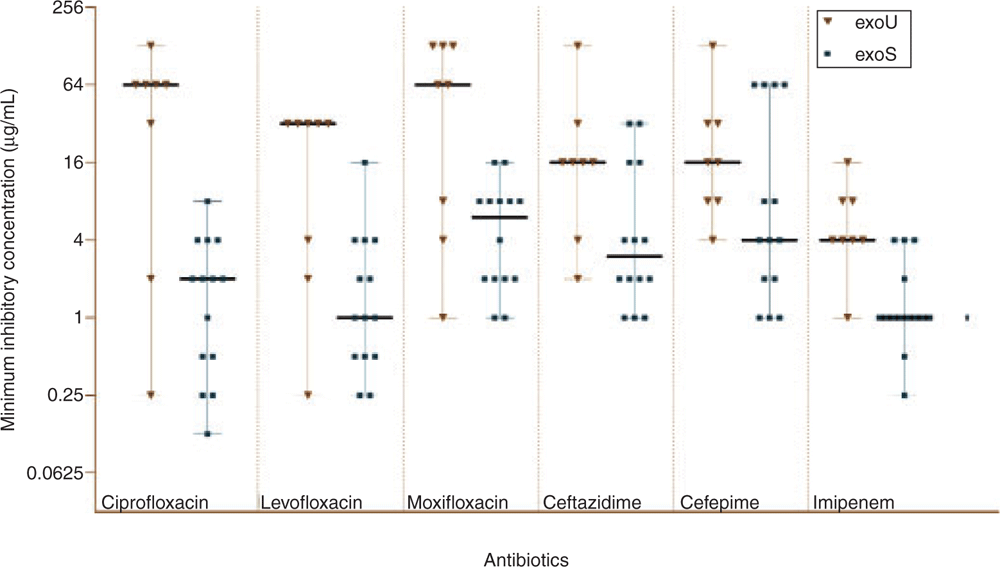
Not only are exoU carrying strains able to cause more severe infections, co-selection of exoU gene and antimicrobial resistance, including disinfectant resistance, has been widely reported in exoU+ ocular and non-ocular strains14,23–26. This has raised concerns that antibiotic resistance may be a factor for the evolution of more virulent strains of P. aeruginosa. Most studies have shown a significant correlation of exoU with fluoroquinolone resistance in P. aeruginosa. The majority of exoU+ strains have mutations in DNA gyrase (gyrA and gyrB) and topoisomerase (parC and parE), which are responsible for fluoroquinolones resistance. However, deletion or acquisition of exoU alone (not the pathogenicity island) in experimental models does not affect fluoroquinolone susceptibility16, which suggests that other genes in the pathogenicity island may be important in the development of this resistance phenotype. Limited studies have also shown a correlation between exoU carriage and resistance to beta-lactams and aminoglycosides16,27. Our previous study examined the correlation between carriage of exoU or exoS and mutation and expression of beta-lactamase genes. We observed that exoU+ strains were usually more resistant to beta-lactams than exoS+ strains (Figure 2)28, and this may have been due to exoU+ strains having more mutations in genes associated with beta-lactam resistance (mexR, ampC and ampR). Gene expression analysis suggested that such mutations generally lead to antibiotic resistance28. However, the reason for a higher mutation rate in above-mentioned resistance genes in exoU+ strains remains unanswered. Perhaps selection pressure, which favours acquisition of exoU+, is associated with higher mutation rates in exoU strains.
|
Taking all the information together, it is clear that exoU in P. aeruginosa is ecologically important and an important determinant of virulence and antimicrobial resistance. Perhaps screening for exoU might help predict clinical outcomes and resistance patterns, which in turn could lead to development of strategies of improved therapies. Furthermore, given that exoU+ strains are distributed differently in the different environment, the exoU profile of strains may help to track the epidemiology of P. aeruginosa infections. Further studies on the incompatibility of exoU and exoS within the same P. aeruginosa genome, and factors favouring acquisition of the exoU might help to understand acquired resistance in P. aeruginosa.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This research did not receive any specific funding.
References
[1] Hauser, A.R. (2009) The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665.| The type III secretion system of Pseudomonas aeruginosa: infection by injection.Crossref | GoogleScholarGoogle Scholar | 19680249PubMed |
[2] Feltman, H. et al. (2001) Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147, 2659–2669.
| Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 11577145PubMed |
[3] Fleiszig, S.M. et al. (1997) Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 65, 579–586.
| 9009316PubMed |
[4] Hauser, A.R. et al. (2002) Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30, 521–528.
| Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 11990909PubMed |
[5] Finck-Barbançon, V. et al. (1997) ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25, 547–557.
| ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury.Crossref | GoogleScholarGoogle Scholar | 9302017PubMed |
[6] Hauser, A.R. et al. (1998) PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27, 807–818.
| PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence.Crossref | GoogleScholarGoogle Scholar | 9515706PubMed |
[7] Wolfgang, M.C. et al. (2003) Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100, 8484–8489.
| Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 12815109PubMed |
[8] Subedi, D. et al. (2018) Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci. Rep. 8, 15668.
| Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites.Crossref | GoogleScholarGoogle Scholar | 30353070PubMed |
[9] Freschi, L. et al. (2018) Genomic characterisation of an international Pseudomonas aeruginosa reference panel indicates that the two major groups draw upon distinct mobile gene pools. FEMS Microbiol. Lett. 365, fny120.
| Genomic characterisation of an international Pseudomonas aeruginosa reference panel indicates that the two major groups draw upon distinct mobile gene pools.Crossref | GoogleScholarGoogle Scholar | 29897457PubMed |
[10] Boulant, T. et al. (2018) Higher prevalence of PldA, a Pseudomonas aeruginosa trans-kingdom H2-type VI secretion system effector, in clinical isolates responsible for acute infections and in multidrug resistant strains. Front. Microbiol. 9, 2578.
| Higher prevalence of PldA, a Pseudomonas aeruginosa trans-kingdom H2-type VI secretion system effector, in clinical isolates responsible for acute infections and in multidrug resistant strains.Crossref | GoogleScholarGoogle Scholar | 30420847PubMed |
[11] Rutherford, V. et al. (2018) Environmental reservoirs for exoS+ and exoU+ strains of Pseudomonas aeruginosa. Environ. Microbiol. Rep. 10, 485–492.
| Environmental reservoirs for exoS+ and exoU+ strains of Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 29687624PubMed |
[12] Bradbury, R.S. et al. (2010) Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 59, 881–890.
| Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 20430902PubMed |
[13] Costerton, J.W. et al. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322.
| Bacterial biofilms: a common cause of persistent infections.Crossref | GoogleScholarGoogle Scholar | 10334980PubMed |
[14] Choy, M.H. et al. (2008) Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis. J. Med. Microbiol. 57, 1539–1546.
| Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis.Crossref | GoogleScholarGoogle Scholar | 19018027PubMed |
[15] Zhu, H. et al. (2006) Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr. Eye Res. 31, 297–306.
| Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis.Crossref | GoogleScholarGoogle Scholar | 16603462PubMed |
[16] Borkar, D.S. et al. (2014) Cytotoxic clinical isolates of Pseudomonas aeruginosa identified during the Steroids for Corneal Ulcers Trial show elevated resistance to fluoroquinolones. BMC Ophthalmol. 14, 54.
| Cytotoxic clinical isolates of Pseudomonas aeruginosa identified during the Steroids for Corneal Ulcers Trial show elevated resistance to fluoroquinolones.Crossref | GoogleScholarGoogle Scholar | 24761794PubMed |
[17] Klockgether, J. et al. (2004) Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa. C. J. Bacteriol. 186, 518.
| Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa. C.Crossref | GoogleScholarGoogle Scholar | 14702321PubMed |
[18] Stover, C.K. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964.
| Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen.Crossref | GoogleScholarGoogle Scholar | 10984043PubMed |
[19] Subedi, D. et al. (2019) Accessory genome of the multi-drug resistant ocular isolate of Pseudomonas aeruginosa PA34. PLoS One 14, e0215038.
| Accessory genome of the multi-drug resistant ocular isolate of Pseudomonas aeruginosa PA34.Crossref | GoogleScholarGoogle Scholar | 30986237PubMed |
[20] Lee, D.G. et al. (2006) Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7, R90.
| Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial.Crossref | GoogleScholarGoogle Scholar | 17038190PubMed |
[21] Kulasekara, B.R. et al. (2006) Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188, 4037–4050.
| Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 16707695PubMed |
[22] Treangen, T.J. et al. (2014) The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524.
| The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes.Crossref | GoogleScholarGoogle Scholar | 25410596PubMed |
[23] Wong-Beringer, A. et al. (2008) Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 14, 330–336.
| Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 18190571PubMed |
[24] Cho, H.H. et al. (2014) Correlation between virulence genotype and fluoroquinolone resistance in carbapenem-resistant Pseudomonas aeruginosa. Ann. Lab. Med. 34, 286–292.
| Correlation between virulence genotype and fluoroquinolone resistance in carbapenem-resistant Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 24982833PubMed |
[25] Sawa, T. et al. (2014) Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review. Crit. Care 18, 668.
| Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review.Crossref | GoogleScholarGoogle Scholar | 25672496PubMed |
[26] Lakkis, C. and Fleiszig, S.M. (2001) Resistance of Pseudomonas aeruginosa isolates to hydrogel contact lens disinfection correlates with cytotoxic activity. J. Clin. Microbiol. 39, 1477–1486.
| Resistance of Pseudomonas aeruginosa isolates to hydrogel contact lens disinfection correlates with cytotoxic activity.Crossref | GoogleScholarGoogle Scholar | 11283074PubMed |
[27] Garey, K.W. et al. (2008) Prevalence of type III secretion protein exoenzymes and antimicrobial susceptibility patterns from bloodstream isolates of patients with Pseudomonas aeruginosa bacteremia. J. Chemother. 20, 714–720.
| Prevalence of type III secretion protein exoenzymes and antimicrobial susceptibility patterns from bloodstream isolates of patients with Pseudomonas aeruginosa bacteremia.Crossref | GoogleScholarGoogle Scholar | 19129069PubMed |
[28] Subedi, D. et al. (2018) Association between possession of ExoU and antibiotic resistance in Pseudomonas aeruginosa. PLoS One 13, e0204936.
| Association between possession of ExoU and antibiotic resistance in Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 30265709PubMed |
Biographies
Dinesh Subedi completed his PhD from the University of New South Wales in the area of antimicrobial resistance. His research interests include molecular microbiology, microbial genomics and antimicrobial resistance. He is a post-doctoral research fellow in Jeremy Barr’s laboratory in the School of Biological Sciences, Monash University.
Ajay Kumar Vijay graduated in 1997 with Bachelor’s Degree in Optometry at the Birla Institute of Technology, after which he worked as a Clinical Optometrist in private practice for 4 years. He completed his doctoral study at the University of New South Wales in 2007, which was focused on developing animal models for infiltrative keratitis and testing antimicrobial contact lenses. After graduating, he worked at the Brien Holden Vision Institute as a Research Associate, conducting laboratory and animal trials investigating antimicrobial compounds as well as managing industry sponsored research projects. Dr Vijay is currently a senior post-doctoral research fellow working with Professor Mark Willcox at the School of Optometry and Vision Science, University of New South Wales. His research areas include contact lenses related infiltrative events, dry eye and antibiotic resistance. Dr Vijay has 26 peer-reviewed publications (h index 8) with several refereed conference abstracts.
Scott A Rice has studied mechanisms of microbial interaction and biofilm development for more than 20 years. After completing a PhD at the University of Tennessee, he moved to Sydney to work on quorum sensing (QS) at the University of New South Wales in the Centre for Marine Bio-Innovation. There, he was involved in both the fundamental aspects of QS control of microbial populations, including virulence factor expression and biofilms as well as developing interference strategies for control of opportunistic pathogens. His research led to discoveries of the role of nitric oxide, nutrient deprivation and bacteriophage in controlling biofilm development. In 2014, he took a full-time position at the Singapore Centre for Environmental Life Sciences Engineering at Nanyang Technological University to focus on mixed species biofilm communities. That work has led to the establishment of various microbial community-based systems to study the ecological interactions of microbes and their consequences for environmental (microbially influenced corrosion, larval settlement), industrial (reverse osmosis membranes) and host systems (skin and gut).
Mark Willcox is a medical microbiologist who has specialised in oral and ocular microbiology in his career. He has particular research interests in the development of new antimicrobials, particularly those that can be coated onto medical devices. His group has recently completed a Phase III clinical trial of an antimicrobial peptide-coated contact lens.


