Variables affecting laboratory diagnosis of acute rickettsial infection
Cecilia KatoRickettsial Zoonoses Branch
Division of Vector-Borne Diseases
National Center For Emerging and Zoonotic Infectious Diseases
Centers for Disease Control and Prevention
MailStop H17-3
1600 Clifton Road NE
Atlanta, GA 30333, USA
Office: 404.639.0152
Email: ckato@cdc.gov
Microbiology Australia 39(4) 220-222 https://doi.org/10.1071/MA18068
Published: 12 November 2018
The reference standard for the confirmation of a recent rickettsial infection is by the observation of a four-fold or greater rise in antibody titres when testing paired acute and convalescent (two to four weeks after illness resolution) sera by serological assays (Figure 1). At the acute stage of illness, diagnosis is performed by molecular detection methods most effectively on DNA extracted from tissue biopsies (eschars, skin rash, and organs) or eschar swabs. Less invasive and more convenient samples such as blood and serum may also be used for detection; however, the low number of circulating bacteria raises the possibility of false negative results. Optimal sampling practices and enhanced sensitivity must therefore be considered in order to provide a more accurate laboratory diagnosis.

|
Human pathogenic bacteria from the genus Rickettsiae cause mild to severe diseases worldwide. The rickettsial agents (spotted fever group) found in Australia include Rickettsia australis, R. honei subsp. marmionii, and R. felis typically cause mild to moderately severe illness. Nonspecific symptoms of all rickettsioses at the early stage of illness confound clinical diagnosis. Patients should be given appropriate antibiotic therapy upon suspicion of having a rickettsial disease because it is essential for effective treatment especially in more severe rickettsioses, such as Rocky Mountain spotted fever (RMSF), where a delay in doxycycline treatment correlates with more dire outcomes and death. RMSF is caused by the Gram-negative Alphaproteobacteria, R. rickettsii. With fatality rates from 5% to 42% in paediatric cases in the US and Mexico1,2, early clinical diagnosis and doxycycline treatment are essential for a positive prognosis. However, clinical diagnosis is difficult because symptoms at the initial stage of illness are nonspecific and may include fever, chills, headache, and malaise; and the characteristic maculopapular rash, which spreads centripetally and can involve soles and palms, may not be seen or may only present later at two to four days after symptom onset2. Although RMSF is not endemic to Australia, international travel and exposure to arthropods should be considered during clinical diagnosis.
Molecular detection is readily reported and may be used for the confirmation of disease at the acute stage of illness. However, because Rickettsia are obligate intracellular bacteria, these organisms localise in endothelial cells and the level circulating in blood is believed to be low at the early stage of infection, less than 100 copies per mL of blood3,4. The low bacteremia may equate to less than 1 genome equivalent per 10 µL of blood. Therefore, rickettsial DNA may not be in the test reaction, or may be present below the reproducible limit of detection5. Positive results confirm disease, but negative results can only describe that there was no detectable target DNA in the reaction. Other factors affecting molecular detection of Rickettsia in blood includes the timing of sample draw, patient antibiotic treatment, sample age, sample stabiliser, and assay sensitivity.
Timing of sample draw and antibiotic treatment
For the detection of rickettsial DNA in blood by molecular methods, the sample must be taken before or within 48 hours of appropriate antibiotic treatment to minimise false-negative results2. Note: antibiotics must not be withheld and patients should be empirically treated upon suspicion of rickettsial infection. Due to the fast progression and potential severity of these diseases, early treatment is essential for the best possible outcome6. False negatives due to low rickettsial bacteremia are difficult to verify so the level of detection efficiency at the acute stage of illness is not clear at this time.
Sample age and blood collection tubes
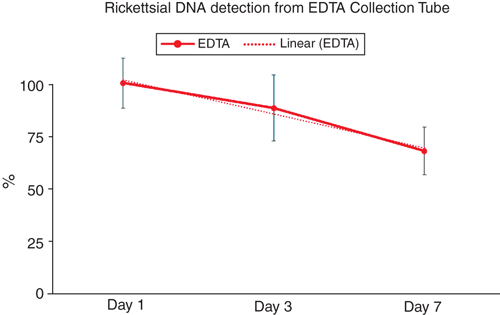
The standard retention time of blood for PCR testing is within seven days of sample draw. Ethylenediaminetetraacetic acid (EDTA) blood collection tubes are used in haematology testing and are reported most often for the molecular detection of Rickettsia2 as well as other infectious diseases. Acid citrate synthase anticoagulants have also been described as acceptable for molecular testing, while heparin has been described as having an inhibitory effect on polymerase activity. The literature describing PCR anticoagulant compatibility originated with protocols using phenol-chloroform extractions and early master mix formulations7. Since these early reports of PCR/ blood stabiliser compatibility, new reaction mix and extraction chemistries and technologies have been developed and must be examined with the reagents and methodologies within individual labs. We verified that the performance of the reagents and extraction products used in our testing are compatible with all three stabilisers stored at 4°C and our current methods8. We examined the level of detection of rickettsial target in contrived samples using blood treated with EDTA, acid citrate dextrose solution A (ACD-A), and sodium heparin over seven days and have verified the use of these blood stabilisation types as compatible with our testing methods. While ACD-A and heparin additives provided testable extraction products comparable to or better than the current collection tube standard, the EDTA samples showed a decline in target detection within the seven day period (Figure 2).
|
Assay sensitivity
Current molecular detection assays for rickettsial diseases include real-time PCR and isothermal amplification protocols with specificities varying from 78% to 99% and limits of detection from one to 10 copies per reaction5. These methodologies are at the limit of detection for these targets and technologies. This calculates to 200 to 2000 genome equivalents per millilitre of blood, which is still above the detection range needed (less than 100 copies per mL of blood) at the early stage of illness. Due to the variation in protocols it is unclear if the differences in sensitivities are due to amplification targets, reagents, instrumentation, extraction methodologies, or assessment strategies. RNA detection has increased the detectable range, as the target numbers may be higher than the DNA copies as long as labile RNA transcripts have not degraded.
Conclusion
There is currently an undefined level of accuracy for molecular detection methods in blood due to current DNA assay sensitivity and overall variation in best practices for sampling, stabilisation, and preparation. It is important to be mindful of the following when testing blood.
-
Draw sample during the symptomatic stage of illness, before or within 48 hours of doxycycline treatment.
-
Samples must be processed as soon as possible or within days to avoid template degradation, especially if EDTA is the blood stabiliser.
-
Assessment of alternative targets might increase assay sensitivity. RNA detection is a promising target and its utility and limitations are yet to be defined.
The optimisation of all preanalytical and analytical processes may improve rickettsial molecular detection in blood at the acute stage of illness. Further validation is needed to determine a standard for sample collection and handling to improve integrity of specimens suspected of rickettsial infection.
Conflicts of interest
The author declares no conflicts of interest.
Acknowledgements
This research did not receive any specific funding.
References
[1] Alvarez-Hernandez, G. et al. (2015) Clinical profile and predictors of fatal Rocky Mountain spotted fever in children from Sonora, Mexico. Pediatr. Infect. Dis. J. 34, 125–130.| Clinical profile and predictors of fatal Rocky Mountain spotted fever in children from Sonora, Mexico.Crossref | GoogleScholarGoogle Scholar |
[2] Biggs, H.M. et al. (2016) Diagnosis and management of tickborne rickettsial diseases: Rocky Mountain spotted fever and other spotted fever group rickettsioses, Ehrlichioses, and Anaplasmosis –United States. MMWR 65, 1–44.
[3] Kaplowitz, L.G. et al. (1983) Correlation of rickettsial titers, circulating endotoxin, and clinical features in Rocky Mountain spotted fever. Arch. Intern. Med. 143, 1149–1151.
| Correlation of rickettsial titers, circulating endotoxin, and clinical features in Rocky Mountain spotted fever.Crossref | GoogleScholarGoogle Scholar |
[4] Kato, C.Y. et al. (2013) Assessment of real-time PCR for detection of Rickettsia spp. and Rickettsia rickettsii in banked clinical samples. J. Clin. Microbiol. 51, 314–317.
| Assessment of real-time PCR for detection of Rickettsia spp. and Rickettsia rickettsii in banked clinical samples.Crossref | GoogleScholarGoogle Scholar |
[5] Paris, D.H. and Dumler, J.S. (2016) State of the art of diagnosis of rickettsial diseases: the use of blood specimens for diagnosis of scrub typhus, spotted fever group rickettsiosis, and murine typhus. Curr. Opin. Infect. Dis. 29, 433–439.
| State of the art of diagnosis of rickettsial diseases: the use of blood specimens for diagnosis of scrub typhus, spotted fever group rickettsiosis, and murine typhus.Crossref | GoogleScholarGoogle Scholar |
[6] Regan, J.J. et al. (2015) Risk factors for fatal outcome from Rocky Mountain spotted fever in a highly endemic area—Arizona, 2002–2011. Clin. Infect. Dis. 60, 1659–1666.
| Risk factors for fatal outcome from Rocky Mountain spotted fever in a highly endemic area—Arizona, 2002–2011.Crossref | GoogleScholarGoogle Scholar |
[7] Holodniy, M. et al. (1991) Inhibition of human immunodeficiency virus gene amplification by heparin. J. Clin. Microbiol. 29, 676–679.
[8] Kato, C. et al. (2016) Estimation of Rickettsia rickettsii copy number in the blood of patients with Rocky Mountain spotted fever suggests cyclic diurnal trends in bacteraemia. Clin. Microbiol. Infect. 22, 394–396.
| Estimation of Rickettsia rickettsii copy number in the blood of patients with Rocky Mountain spotted fever suggests cyclic diurnal trends in bacteraemia.Crossref | GoogleScholarGoogle Scholar |
Biography
Dr Cecilia Kato is the Rickettsia Diagnostics Team Lead and Director of the Reference Diagnostic Laboratory at the Centers for Disease Control and Prevention, Division of Vector Borne Diseases, Rickettsial Zoonoses Branch in Atlanta Georgia, in the United States. Cecilia has been at the CDC since 2008 and has served as the lead of the Diagnostics Lab since 2012. The Rickettsia Diagnostics Team supports United States and international Public Health Labs with the laboratory diagnosis of rickettsiosis, ehrlichiosis, anaplasmosis, scrub typhus, and Q fever. Her research emphasis is on assay development, which includes a provisional patent for enhanced sensitivity for Rickettsia species detection in patient samples, a FDA cleared Rickettsia real-time PCR kit, point of care diagnostics, and enhanced surveillance. She also works with international partners to build diagnostic capacity and supports epidemiological studies and outbreak response.


