Candida auris: the most talked about multidrug-resistant emerging fungal pathogen
Laszlo Irinyi A B , Richard Malik C and Wieland Meyer A B *A Curtin Medical School, Faculty of Health Sciences, Curtin University, Kent Street, Perth, WA 6102, Australia.
B Westmead Clinical School, Sydney Medical School, Faculty of Medicine and Health, The University of Sydney, Sydney, NSW 2006, Australia.
C Centre for Veterinary Education, The University of Sydney, Camperdown, NSW 2006, Australia.

Dr Laszlo Irinyi is a Research Fellow at Curtin University, WA, Australia and The University of Sydney, Sydney, NSW, Australia. His research centres around the adaptation of next generation sequencing technologies in precision and real-time diagnostic of fungal diseases. In addition, he is interested in leveraging big data archived in public databases for the search of the origin and spread of emerging pathogens, as well as to assess the fungal diversity in various environments using metabarcoding/metagenomic approaches. |

Dr Richard Malik is a veterinarian with a strong interest in infectious diseases, especially fungi and parasites. He works for the Centre for Veterinary Education at the University of Sydney. |

Professor Wieland Meyer is a Molecular Medical Mycologist and academic at the Curtin Medical School, Faculty of Health Science, Curtin University; Sydney Medical School, Faculty of Medicine and Health, The University of Sydney, Sydney, NSW, Australia; and the Fundação Oswaldo Cruz (FIOCRUZ), Rio de Janeiro, Brazil. His research focuses on phylogeny, molecular identification, population genetics, molecular epidemiology, and virulence mechanisms of human and animal pathogenic fungi. He is the Convener of the Mycology Interest Group of ASM, and the President of the International Mycological Association (IMA). |
Microbiology Australia 43(4) 173-176 https://doi.org/10.1071/MA22057
Submitted: 7 November 2022 Accepted: 7 December 2022 Published: 23 December 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Currently Candida auris is the most talked about multidrug-resistant emerging fungal pathogen. It can cause difficult-to-control nosocomial outbreaks worldwide, being highly contagious, and poses serious challenges to public health authorities. A wide spectrum of infections, ranging from superficial mucosal infections, candidemia to disseminated deep-seated disease, having been reported from more than 40 countries, including Australia. Outbreaks are associated with high mortality rates due to rapid transmission and challenges in prevention, control, and treatment. It is shows a high multidrug-resistance (with 90% of isolates resistant to fluconazole), and is extremely tolerant to conditions which usually limit fungal transmission, including commonly used disinfectants. Whole genome analysis has revealed five different closely related clades (named after the geographical areas they have been first encoundered) with distinct clonal lineages. Its environmental niche remained a mystery until recently when it was found in India in association with costal aquatic environments.
Keywords: alternative treament options, Australian situation, Candida auris, clinical issues, emerging fungal pathogens, environmental sources, laboratory identification, multidrug resistance.
Introduction
Candida species are the most common opportunistic fungal pathogens in immunosuppressed patients as a result of treatment of cancer or auto-immune diseases with chemotherapy and/or immunotherapy.1 Candida spp. are found on the skin, in the oral cavity, gastrointestinal, and urogenital tract of healthy individuals, where they colonise these surfaces as commensal microorganisms. When there is a shift in the host–microbial relationship, they proliferate and sometimes invade, giving rise to vaginitis, oral and oesophageal candidiasis, cutaneous candidiasis, candidemia, and systemic infections.1 Among them, candidemia (bloodstream infection) is the most frequent hospital-acquired fungal infection. Invasive Candida infections are associated with high rates of morbidity and mortality, increased length of hospital stay, and hugely elevated healthcare costs.2 Increasing populations of immunocompromised people due to invasive clinical procedures, organ transplantation, cancer treatment, management of immune-mediated diseases, HIV infection/AIDS, and long periods of hospitalisation contribute to high invasive fungal disease rates.3
Candida auris
C. auris is an emerging multidrug-resistant yeast in the Candida/Clavispora clade of the family Metschnikowiaceae. It is primarily a skin and mucous membrane commensal but is capable of causing invasive fungal disease and difficult-to-control outbreaks in healthcare-associated settings globally, often in intensive care units (ICUs).4–6 Phylogenetically, it is closely related to the Candida haemulonii species complex (C. haemulonii, C. haemulonii var. vulnera, C. duobushaemulonii, C. pseudohaemulonii, and C. vulturna), C. heveicola, and Candida ruelliae, from which it can be separated by sequencing the internal transcribed spacer (ITS) regions of the nuclear rRNA gene operon (Fig. 1).7 C. auris isolates are often resistant or even multi-resistant to antifungal drugs and are easily transmitted from person to person in healthcare settings, so its recent emergence causing major outbreaks globally has caused significant public health concerns.8,9
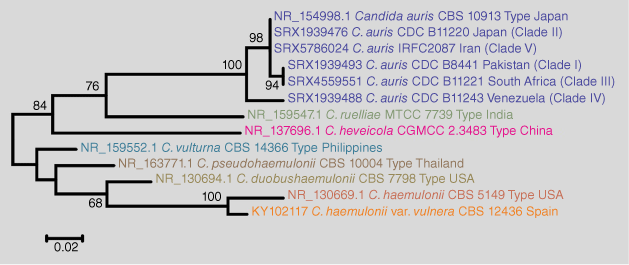
|
Genetic diversity
Isolates of C. auris have been classified in five distinct clades named after the geographical areas where they were firstly described (South Asian (Clade 1), East Asian (Clade 2), African (Clade 3), South American (Clade 4) and an Iranian (Clade 5)), harbouring nearly identical strains within each clade but show thousands of single nucleotide polymorphism (SNP) differences between the clades based on whole genome sequencing.10,11 The low level of genetic diversity within the clades and the high diversity between the clades suggests that C. auris evolved at least 360 years ago when applying a Bayesian approach for molecular dating.12
Environmental sources
There are ongoing speculations that the sudden emergence of resistant or multi-resistant yeast species is due to one or a combination of the following factors: (1) widespread use of antifungal agents in agriculture; (2) climate change, which enables them to grow at mammalian body temperature, becoming pathogens and causing disease in humans; (3) the role of healthcare settings supporting the transmission of these species within and across their networks; (4) possible changes in the organisms by sharing virulence and drug-resistance characteristics within the C. haemulonii clade; (5) recently increased human contacts with unknown ecological niches such as plant, insect, soil, or aquatic habitats; and (6) large-scale environmental changes including expansion of industrial farming practices, deforestation, agricultural expansion, coastal ecosystem disruption enhancing the amplification of these species.8,11,13,14 In addition, it was proposed that C. auris might have intermediate hosts, such as avian species, which facilitate its spread and indirect transmission to humans from the environment.14
Based on whole genome sequencing and epidemiologic analyses, C. auris appeared to emerge nearly simultaneously on three continents and has since spread across the globe.10 Its precise origin and true environmental niches remain a mystery. C. auris was isolated from two indoor swimming pool facilities from The Netherlands indicating potential transmission pathways of the ‘superbug fungus’.15 The first environmental isolates of C. auris were obtained only very recently from the Coastal Wetlands of the Andaman Islands, India.16 A recent in silico species-specific sequence search in the world’s largest sequence repository, the Sequence Read Archive (SRA) at NCBI identified potential environmental sources of C. auris such as airborne dust, activated sludge, peanut fields, and faeces.17 These results suggest a possible ecological niche of C. auris in wetlands where it can be associated with amphibians, from where it can spread via water, aerosols or bird species to human or animal habitats. The same study concluded that C. auris could infect other animals such as dogs and amphibians beside humans.17 In addition, it has been shown that the life cycle could be completed by excretion of C. auris from infected humans/animals via faeces, which then contaminate wastewater, in which it is able to survive despite wastewater treatment (sludge) prior to returning to wetlands.17
Clinical issues
C. auris was originally described from an ear infection in a Japanese patient in 2009.7 However, it had first been isolated in 1996 from blood of a patient in a South Korean hospital, but having been misidentified as C. haemulonii.18 Another 15 C. haemulonii isolates from ear canals of patients with chronic otitis at five university hospitals in Korea were later identified to be C. auris.19
Since its first description, C. auris has been reported to cause serious invasive blood stream infections with a high mortality (~50%), especially in patients in hospital ICUs.4,20,21 The first three cases of nosocomial fungemia caused by C. auris were reported from South Korea in 2011.21 Since then, it has been identified as the cause of serious infections in every continent (except Antarctica) and in a variety of clinical settings.6 To date C. auris infections and associated outbreaks have been reported in more than 40 countries on six continents.5,12
C. auris is able to persist in the hospital environment and on the surface of medical equipment and has developed resistance to disinfectants commonly used in daily practice.22 In vitro survival studies found that C. auris can survive for up to 1 month on different abiotic substrates, such as plastic and steel.23 It has the capacity to form antifungal resistant biofilms sensitive to the disinfectant chlorhexidine in vitro24 and its virulence mechanisms enables it to evade the immune response or help to avoid the attack of neutrophils.25 Moreover, C. auris can easily colonise the skin, allowing for the transmission from person-to-person through direct contact.4
Resistance, possible alternative treatments and virulence
C. auris is reported to be resistant to multiple antifungal drugs: 90% of isolates are fluconazole resistant and many isolates are cross-resistant to more than one of the three major classes of antifungals – azoles, echinocandins and polyenes. The development of antifungal resistance could be due to the excessive use of antifungals in the agricultural sector. The mechanisms for antifungal resistance are still not clear.10 Given the high resistance in a number of C. auris strains, new strategies for treatment need to be explored. One possibility is to take advantage of synergistic effects between certain antifungals, with the highest synergism found between voriconazole and micafungin (100%), flucytosine and amphotericin B and flucytosine and micafungin (both 7%) and another possibility is to explore synergisms between antifungals and antibiotics, for example, caspofungin and colistin (100%). Both approaches have currently only been experimentally tested.26
Recent comparative genomics studies showed that C. auris carries extensive mutations and copy number variations in genes linked to several virulence factors, such as lipases, phospholipase and proteinases, or enabling biofilm formation and adherence to plastic surfaces.24
Laboratory identification
Commercial systems routinely used in clinical laboratories for yeast identification such as VITEK (BioMerieux) and API-20C AUX (BioMerieux) often misidentify (up to 90%) the pathogen as C. haemulonii and Rhodotorula glutinis, respectively,21 due to the lack of C. auris profiles in their databases. The real prevalence of C. auris infections may therefore be underestimated.6 If the right reference spectral database is in place Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-ToF) can readily differentiate C. auris from other Candida species. On traditional CHROMagar™ C. auris has a smooth pale pinkish colony appearance, making it somehow hard to be differentiated from C. glabrata (dark violet), Pichia kudriavzevii (formerly C. krusei) (pinkish, flat, rough, pale border) and Meyerozyma guilliermondii (formerly C. guilliermondii) (pink/lavender, mucoid). The recently developed CHROMagar™ Candida Plus selective chromogenic culture medium permits the rapid identification of C. auris and reliably differentiates it from other common Candida species, based on its white/light blue creamy colonies with a blue halo appearance (see cover photo, left C. auris on CHROMagar™ right C. auris on CHROMagar™ Candida Plus). Recently a real-time PCR protocol was developed in an Australian clinical diagnostic setting for the identification of C. auris in skin swabs for high-throughput infection control screening.27
The Australian situation
In 2015, C. auris was reported for the first time in Australia as a cause of sternal osteomyelitis in a man visiting Western Australia. The patient had a history of intensive care treatment in Kenya.28 Later in 2010, four patients with previous overseas hospitalisation were identified with C. auris in Victoria.29 Phylogenetic analysis revealed presumed transmission between two of these patients and suspected overseas acquisition in the others. In 2020, a study described the genetic heterogeneity of Australian C. auris isolates from a non-outbreak setting in Sydney hospitals using whole genome sequencing.30 The isolates were genetically distinct, with no isolate being identical to another, and multidrug-resistance was absent.
Conclusion
The global spread of C. auris, resulting in its recent listing as the second major human fungal pathogen on the first official WHO Fungal Pathogen Priority List (WHO),31 highlights the importance of raising awareness of the increasing risk, that travellers coming to Australia from highly endemic C. auris regions (such as the Indian subcontinent) pose in bringing the pathogen with them into Australian health care settings. Overall, the cases seen in the Australian hospital settings have been in travellers returning to Australia, where they either became colonised or infected during hospitalisation overseas. It also emphasises the need to establish appropriate diagnostic procedures and implementation of local clinical surveillance accompanied by genomic monitoring to enable an early detection and an effective public health response if needed.
Data availability
All data are available from Genbank.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This study was supported by a National Health and Medical Research Council of Australia (NHMRC) grant [no. APP1121936] to Wieland Meyer.
References
[1] Pfaller, MA et al.. (2014) Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004–2008. PLoS ONE 9, e101510.| Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004–2008.Crossref | GoogleScholarGoogle Scholar |
[2] Benedict, K et al.. (2019) Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis 68, 1791–7.
| Estimation of direct healthcare costs of fungal diseases in the United States.Crossref | GoogleScholarGoogle Scholar |
[3] Sabino, R et al.. (2020) Candida auris, an agent of hospital-associated outbreaks: which challenging issues do we need to have in mind? Microorganisms 8, 181.
| Candida auris, an agent of hospital-associated outbreaks: which challenging issues do we need to have in mind?Crossref | GoogleScholarGoogle Scholar |
[4] Chowdhary, A et al.. (2017) Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13, e1006290.
| Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally.Crossref | GoogleScholarGoogle Scholar |
[5] Saris, K et al.. (2018) Candida auris. Curr Opin Infect Dis 31, 334–40.
| Candida auris.Crossref | GoogleScholarGoogle Scholar |
[6] Du, H et al.. (2020) Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog 16, e1008921.
| Candida auris: epidemiology, biology, antifungal resistance, and virulence.Crossref | GoogleScholarGoogle Scholar |
[7] Satoh, K et al.. (2009) Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53, 41–4.
| Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital.Crossref | GoogleScholarGoogle Scholar |
[8] Jackson, BR et al.. (2019) On the origins of a species: what might explain the rise of Candida auris? J Fungi (Basel, Switzerland) 5, 58.
| On the origins of a species: what might explain the rise of Candida auris?Crossref | GoogleScholarGoogle Scholar |
[9] Gade, L et al.. (2020) Understanding the emergence of multidrug-resistant Candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet 11, 554.
| Understanding the emergence of multidrug-resistant Candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex.Crossref | GoogleScholarGoogle Scholar |
[10] Lockhart, SR et al.. (2017) Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64, 134–40.
| Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses.Crossref | GoogleScholarGoogle Scholar |
[11] Chow, NA et al.. (2018) Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18, 1377–84.
| Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey.Crossref | GoogleScholarGoogle Scholar |
[12] Chow, NA et al.. (2020) Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 11, e03364-19.
| Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses.Crossref | GoogleScholarGoogle Scholar |
[13] Muñoz, JF et al.. (2018) Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 9, 5346.
| Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species.Crossref | GoogleScholarGoogle Scholar |
[14] Casadevall, A et al.. (2021) Environmental Candida auris and the global warming emergence hypothesis. mBio 12, e00360-21.
| Environmental Candida auris and the global warming emergence hypothesis.Crossref | GoogleScholarGoogle Scholar |
[15] Ekowati, Y et al.. (2018) Potential transmission pathways of clinically relevant fungi in indoor swimming pool facilities. Int J Hyg Environ Health 221, 1107–15.
| Potential transmission pathways of clinically relevant fungi in indoor swimming pool facilities.Crossref | GoogleScholarGoogle Scholar |
[16] Arora, P et al.. (2021) Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio 12, e03181-20.
| Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India.Crossref | GoogleScholarGoogle Scholar |
[17] Irinyi, L et al.. (2022) Finding a needle in a haystack – in silico search for environmental traces of Candida auris. Jpn J Infect Dis 75, 490–5.
| Finding a needle in a haystack – in silico search for environmental traces of Candida auris.Crossref | GoogleScholarGoogle Scholar |
[18] Kwon, YJ et al.. (2019) Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol 57, e01624-18.
| Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping.Crossref | GoogleScholarGoogle Scholar |
[19] Kim, MN et al.. (2009) Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48, e57–61.
| Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features.Crossref | GoogleScholarGoogle Scholar |
[20] Schelenz, S et al.. (2016) First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5, 35.
| First hospital outbreak of the globally emerging Candida auris in a European hospital.Crossref | GoogleScholarGoogle Scholar |
[21] Lee, WG et al.. (2011) First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49, 3139–42.
| First three reported cases of nosocomial fungemia caused by Candida auris.Crossref | GoogleScholarGoogle Scholar |
[22] Ku, TSN et al.. (2018) Candida auris: disinfectants and implications for infection control. Front Microbiol 9, 726.
| Candida auris: disinfectants and implications for infection control.Crossref | GoogleScholarGoogle Scholar |
[23] Welsh, RM et al.. (2017) Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55, 2996–3005.
| Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface.Crossref | GoogleScholarGoogle Scholar |
[24] Sherry, L et al.. (2017) Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis 23, 328–31.
| Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris.Crossref | GoogleScholarGoogle Scholar |
[25] Johnson, CJ et al.. (2018) Emerging fungal pathogen Candida auris evades neutrophil attack. mBio 9, e01403-18.
| Emerging fungal pathogen Candida auris evades neutrophil attack.Crossref | GoogleScholarGoogle Scholar |
[26] Aghaei Gharehbolagh, S et al.. (2021) New weapons to fight a new enemy: a systematic review of drug combinations against the drug-resistant fungus Candida auris. Mycoses 64, 1308–16.
| New weapons to fight a new enemy: a systematic review of drug combinations against the drug-resistant fungus Candida auris.Crossref | GoogleScholarGoogle Scholar |
[27] Crawford, LC et al.. (2022) Candida auris PCR for high-throughput infection control screening. Med Mycol 60, myac057.
| Candida auris PCR for high-throughput infection control screening.Crossref | GoogleScholarGoogle Scholar |
[28] Heath, CH et al.. (2019) Candida auris sternal osteomyelitis in a man from Kenya visiting Australia, 2015. Emerg Infect Dis 25, 192–4.
| Candida auris sternal osteomyelitis in a man from Kenya visiting Australia, 2015.Crossref | GoogleScholarGoogle Scholar |
[29] Lane, CR et al.. (2020) Incursions of Candida auris into Australia, 2018. Emerg Infect Dis 26, 1326–8.
| Incursions of Candida auris into Australia, 2018.Crossref | GoogleScholarGoogle Scholar |
[30] Biswas, C et al.. (2020) Genetic heterogeneity of Australian Candida auris isolates: insights from a nonoutbreak setting using whole-genome sequencing. Open Forum Infect Dis 7, ofaa158.
| Genetic heterogeneity of Australian Candida auris isolates: insights from a nonoutbreak setting using whole-genome sequencing.Crossref | GoogleScholarGoogle Scholar |
[31] World Health Organization (WHO) (2022) WHO fungal priority pathogens list to guide research, development and public health action. World Health Organization, Geneva.


