Moonlighting in the matrix
Cynthia B. Whitchurch A B *A Quadram Institute Bioscience, Norwich Research Park, Norwich, NR4 7UQ, UK.
B School of Biological Sciences, University of East Anglia, Norwich, NR4 7TJ, UK.

Cynthia Whitchurch FAA is a Professor of Microbiology at the Quadram Institute Bioscience. Her research interests are in alternate bacterial lifestyles including biofilms and cell wall deficiency. |
Microbiology Australia 44(2) 75-78 https://doi.org/10.1071/MA23022
Submitted: 20 April 2023 Accepted: 11 May 2023 Published: 25 May 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
The biofilm matrix is a complex, heterogeneous mixture of polymers, macromolecules, small molecules, and higher-order particulate structures. The primary functions of some matrix components are specific for building biofilms. However, there are many components of the biofilm matrix that have primary cellular functions yet provide additional distinct ‘moonlighting’ functions when located in the biofilm matrix. Moonlighting matrix components include proteins and nucleic acids as well as higher-order structures such as membranes and bacteriophage. This review will describe some of the moonlighting matrix components found in Pseudomonas aeruginosa biofilms.
Keywords: bacteriophage, biofilm, BMV, EPC, EPS, eDNA, extracellular DNA, MV, OMV, Pseudomonas, slime.
Introduction
Biofilms are multicellular microbial communities or populations that are held together by a complex matrix comprised of extracellular polymeric substances (EPS). The biofilm matrix provides a variety of functions including attachment, aggregation, structural integrity, protection from environmental stresses, antibiotics and disinfectants, facilitation of collective behaviours, provision of nutrients, interaction with host cells and tissues, and contribution to pathogenicity. The biofilm matrix is a complex, heterogeneous mixture of water, large polymers (e.g. exopolysaccharides, DNA, RNA), macromolecules (e.g. proteins, lipids), small molecules (e.g. nucleotides, quorum sensing molecules, siderophores, phenazines) and higher-order particulate structures (e.g. membrane vesicles or MVs, pili, flagella, bacteriophage, cellular debris).1
Some matrix constituents are specifically produced by bacteria for the purposes of biofilm development. Examples of these in the Pseudomonas aeruginosa biofilm matrix are the exopolysaccharides Pel and Psl and the fibrillar adhesin CdrA.2 However, some biofilm matrix components have primary cytosolic functions but can provide other distinct ‘moonlighting’ functions when located in the biofilm matrix. There are many examples of moonlighting components found in the matrix of biofilms of different bacterial species. This paper focuses on moonlighting matrix components found in P. aeruginosa biofilms.
The concept of biological molecules performing moonlighting roles has classically been applied to moonlighting proteins in which a single polypeptide chain can perform two or more distinct functions.3 There are hundreds of examples of moonlighting proteins from animals, plants, yeast and bacteria.3 The majority of bacterial moonlighting proteins are cytosolic, housekeeping proteins that when situated outside of the cell provide moonlighting roles as virulence factors and adhesins.4 As cytosolic moonlighting proteins lack signal peptides, the mechanism by which they reach the cell surface or extracellular milieu is a hotly debated topic.4 Bacterial excretion of cytoplasmic proteins is often associated with factors that affect cell wall structure leading to cell lysis or membrane blebbing. However, some moonlighting proteins may be transported by as yet unidentified excretion routes.4
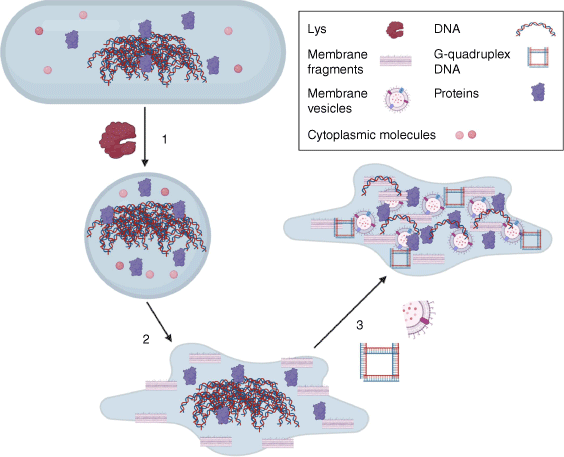
We have shown recently that, in P. aeruginosa, endolysin-mediated explosive cell lysis releases extracellular DNA (eDNA), which is required for biofilm development5,6 (Fig. 1). This autolytic programmed cell-death mechanism also releases cytosolic content and cellular debris into the biofilm extracellular milieu and is the primary mechanism by which MVs are produced in P. aeruginosa biofilms.5 We found that P. aeruginosa mutants that lack the endolysin Lys or the holins Hol, AlpB and CidAB are defective in the initiation and consolidation of biofilm microcolonies.5,7 Interestingly, although eDNA is necessary for biofilm formation by P. aeruginosa,6 it is not sufficient,5,8 which suggests that at least some cellular components released through explosive cell lysis have roles in P. aeruginosa biofilms. The mere presence of cellular components in the biofilm matrix isn’t sufficient to indicate that they provide a function. However, it is now evident that many do moonlight in the matrix.
Explosive cell lysis releases moonlighting matrix materials. (1) The endolysin Lys hydrolyses peptidoglycan and is required for the transition to a spherical morphotype, which (2) then undergoes explosive cell lysis to release intracellular contents and cellular debris; (3) Membrane fragments vesicularise into MVs capturing cellular cargo and eDNA can adopt a G-quadruplex structure. Figure generated with Biorender.com.
Moonlighting matrix proteins
Analysis of the biofilm matrix proteome of P. aeruginosa shows the presence of a number of cytoplasmic proteins.9,10 It has been speculated that positively charged cytoplasmic proteins present in the biofilm matrix of Staphylococcus aureus contribute to biofilm formation through electrostatic interactions with eDNA forming an electrostatic net that binds protein-coated cells to eDNA.11 It is therefore likely that matrix-located cytoplasmic proteins of P. aeruginosa provide similar moonlighting functions. A number of the cytosolic proteins found in the P. aeruginosa biofilm matrix have also been reported to have moonlighting activities involved in host cell interactions in P. aeruginosa and other bacteria. These moonlighting functions include binding to host proteins and mucins by the glycolytic enzyme GAPDH, the chaperones GroEL (HSP60) and DnaK (HSP70), and the protein elongation factor Ef-TU.12 The redox protein azurin involved in intracellular electron-transport chain moonlights extracellularly as a toxin that induces apoptosis of host cells.13 Although these moonlighting functions of proteins in the biofilm matrix have not been directly demonstrated, it is conceivable that these proteins in the biofilm matrix could facilitate attachment to host cells and tissues and contribute to P. aeruginosa virulence.
Other potential moonlighting proteins found in the biofilm matrix of a number of bacterial species including P. aeruginosa are the DNABII nucleoid-binding proteins IHF (integration host factor) and HU (histone-like protein U).14 When located in the cytoplasm, these nucleoid-associated proteins (NAPs) are involved in the maintenance of the structure and function of bacterial chromatin and show an affinity for bent and forked DNA structures. When in the biofilm matrix, these NAPs associate with eDNA and are involved in maintaining the forked network structure of eDNA. Treatment of biofilms with antibodies to these DNABII proteins leads to dissolution of the biofilm indicating their importance in maintaining the structural integrity of the biofilm matrix.14
Extracellular DNA is a moonlighting matrix molecule
Proteins are not the only biological molecules that can have moonlighting roles. The primary function of DNA is the storage of genetic information. However, it is now evident that eDNA is a functional component of the biofilm matrix of P. aeruginosa with roles unrelated to its ability to encode genetic information. Therefore, eDNA can also be considered a moonlighting molecule.
Extracellular DNA was first shown to be required for biofilm formation in P. aeruginosa6 and is now recognised to be an almost universal constituent of the biofilm matrix of many species of bacteria and fungi.15 DNA is a highly versatile molecule that provides a range of moonlighting functions when present in the matrix of P. aeruginosa biofilms including as a ‘glue’ for intercellular connectivity and attachment to the substratum6,16; contribution to biofilm viscoelasticity17; utilisation as a nutrient source under starvation conditions18,19; cation chelation and resistance to antibiotics and antimicrobial peptides through sequestration, acidification and induction of resistance mechanisms20–22; and extracellular electron transport through intercalation of pyocyanin.23,24 In P. aeruginosa, eDNA also contributes to the self-organisation of complex collective behaviours during active biofilm expansion by twitching motility,25 and maturation of submerged, sessile biofilms.26 The structure of eDNA in the extracellular matrix of P. aeruginosa biofilms differs from that of chromosomal DNA within the cell. The higher-order network structure of eDNA in the biofilm matrix contains G-quadruplex structures produced through non-canonical base-pairing. This unusual structure has been proposed to account for the viscoelasticity of eDNA and its role as a primary structural component of the matrix.17
Higher-order structures that moonlight in the matrix
The concept of moonlighting may also be extended to higher-order structures found in the matrix of P. aeruginosa biofilms. For instance, the primary function of bacterial membranes is to act as a selective-permeability barrier and sequester the molecules of life, separating them from the environment. However, membrane fragments and MVs are common components of the P. aeruginosa biofilm matrix.5,27 In this context, membranes are thought to interact directly with eDNA and provide moonlighting matrix functions including contributing to structural integrity, sequestering antibiotics and participating in horizontal gene transfer by natural transformation.27–29
The primary functions of bacteriophage are to infect and replicate in prey bacteria. However, it is now apparent that bacteriophage may also have moonlighting roles in the biofilm matrix. The filamentous Pf bacteriophage of P. aeruginosa promotes self-assembly of biofilm matrix polymers into liquid crystalline structures that enhance biofilm adhesion, viscosity and tolerance to desiccation and antibiotics.30
Concluding remarks
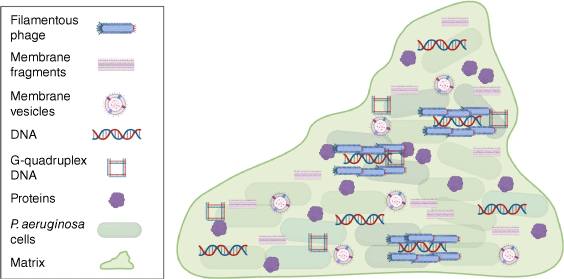
Over 20 years have passed since eDNA was shown to be required for biofilm formation by P. aeruginosa.6 It is now evident that there are many molecules and higher-order structures in the P. aeruginosa biofilm matrix that have key moonlighting roles (Fig. 2). Moonlighting in the matrix is a phenomenon that is certainly not unique to P. aeruginosa biofilms. For instance, moonlighting eDNA is a common component of the matrix of many bacterial biofilms15 and moonlighting cytosolic proteins have been found in the biofilm matrix of several bacterial species.4 It is now clear that biofilm matrixomes contain many cell-derived molecules that may provide moonlighting functions. We do not yet have a complete understanding of how many of the moonlighting matrix components are excreted and evidence exists for both lysis or more specialised secretion systems. It is becoming increasingly evident that cytosolic components present in the biofilm matrix should not be simply overlooked as artefacts of cell lysis. However, determining which of these provide true moonlighting functions and are not just inert passengers in the matrix poses a significant challenge.
Moonlight matrix materials. The P. aeruginosa biofilm matrix contains a number of moonlighting materials including proteins, extracellular DNA, filamentous phage, MVs and membrane fragments. Figure generated with Biorender.com.
References
[1] Flemming, H-C et al. (2023) The biofilm matrix: multitasking in a shared space. Nat Rev Microbiol 21, 70–86.| The biofilm matrix: multitasking in a shared space.Crossref | GoogleScholarGoogle Scholar |
[2] Reichhardt, C (2023) The Pseudomonas aeruginosa biofilm matrix protein CdrA has similarities to other fibrillar adhesin proteins. J Bacteriol , e00019-23.
| The Pseudomonas aeruginosa biofilm matrix protein CdrA has similarities to other fibrillar adhesin proteins.Crossref | GoogleScholarGoogle Scholar |
[3] Jeffery, CJ (2014) An introduction to protein moonlighting. Biochem Soc Trans 42, 1679–1683.
| An introduction to protein moonlighting.Crossref | GoogleScholarGoogle Scholar |
[4] Ebner, P and Götz, F (2019) Bacterial excretion of cytoplasmic proteins (ECP): occurrence, mechanism, and function. Trends Microbiol 27, 176–187.
| Bacterial excretion of cytoplasmic proteins (ECP): occurrence, mechanism, and function.Crossref | GoogleScholarGoogle Scholar |
[5] Turnbull, L et al. (2016) Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7, 11220.
| Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms.Crossref | GoogleScholarGoogle Scholar |
[6] Whitchurch, CB et al. (2002) Extracellular DNA required for bacterial biofilm formation. Science 295, 1487.
| Extracellular DNA required for bacterial biofilm formation.Crossref | GoogleScholarGoogle Scholar |
[7] Hynen, AL et al. (2021) Multiple holins contribute to extracellular DNA release in Pseudomonas aeruginosa biofilms. Microbiology 167, 000990.
| Multiple holins contribute to extracellular DNA release in Pseudomonas aeruginosa biofilms.Crossref | GoogleScholarGoogle Scholar |
[8] Pingle, H et al. (2018) Minimal attachment of Pseudomonas aeruginosa to DNA modified surfaces. Biointerphases 13, 06E405.
| Minimal attachment of Pseudomonas aeruginosa to DNA modified surfaces.Crossref | GoogleScholarGoogle Scholar |
[9] Toyofuku, M et al. (2012) Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res 11, 4906–4915.
| Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix.Crossref | GoogleScholarGoogle Scholar |
[10] Couto, N et al. (2015) Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J Proteome Res 14, 4207–4222.
| Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms.Crossref | GoogleScholarGoogle Scholar |
[11] Dengler, V et al. (2015) An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J Bacteriol 197, 3779–3787.
| An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus.Crossref | GoogleScholarGoogle Scholar |
[12] Jeffery, CJ (2018) Protein moonlighting: what is it, and why is it important? Phil Trans R Soc B 373, 20160523.
| Protein moonlighting: what is it, and why is it important?Crossref | GoogleScholarGoogle Scholar |
[13] Goto, M et al. (2003) Induction of apoptosis in macrophages by Pseudomonas aeruginosa azurin: tumour-suppressor protein p53 and reactive oxygen species, but not redox activity, as critical elements in cytotoxicity. Mol Microbiol 47, 549–559.
| Induction of apoptosis in macrophages by Pseudomonas aeruginosa azurin: tumour-suppressor protein p53 and reactive oxygen species, but not redox activity, as critical elements in cytotoxicity.Crossref | GoogleScholarGoogle Scholar |
[14] Novotny, LA et al. (2016) Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. eBioMedicine 10, 33–44.
| Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo.Crossref | GoogleScholarGoogle Scholar |
[15] Campoccia, D et al. (2021) Extracellular DNA (eDNA). A major ubiquitous element of the bacterial biofilm architecture. Int J Mol Sci 22, 9100.
| Extracellular DNA (eDNA). A major ubiquitous element of the bacterial biofilm architecture.Crossref | GoogleScholarGoogle Scholar |
[16] Das, T et al. (2011) DNA-mediated bacterial aggregation is dictated by acid–base interactions. Soft Matter 7, 2927–2935.
| DNA-mediated bacterial aggregation is dictated by acid–base interactions.Crossref | GoogleScholarGoogle Scholar |
[17] Seviour, T et al. (2021) The biofilm matrix scaffold of Pseudomonas aeruginosa contains G-quadruplex extracellular DNA structures. NPJ Biofilms Microbiomes 7, 27.
| The biofilm matrix scaffold of Pseudomonas aeruginosa contains G-quadruplex extracellular DNA structures.Crossref | GoogleScholarGoogle Scholar |
[18] Finkel, SE and Kolter, R (2001) DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol 183, 6288–6293.
| DNA as a nutrient: novel role for bacterial competence gene homologs.Crossref | GoogleScholarGoogle Scholar |
[19] Mulcahy, H et al. (2010) Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol 12, 1621–1629.
| Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source.Crossref | GoogleScholarGoogle Scholar |
[20] Mulcahy, H et al. (2008) Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4, e1000213.
| Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms.Crossref | GoogleScholarGoogle Scholar |
[21] Lewenza, S (2013) Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front Microbiol 4, 21.
| Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar |
[22] Wilton, M et al. (2016) Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 60, 544–553.
| Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar |
[23] Das, T et al. (2015) Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci Rep 5, 8398.
| Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation.Crossref | GoogleScholarGoogle Scholar |
[24] Saunders, SH et al. (2020) Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 182, 919–932.e19.
| Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms.Crossref | GoogleScholarGoogle Scholar |
[25] Gloag, ES et al. (2013) Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA 110, 11541–11546.
| Self-organization of bacterial biofilms is facilitated by extracellular DNA.Crossref | GoogleScholarGoogle Scholar |
[26] Barken, KB et al. (2008) Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 10, 2331–2343.
| Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms.Crossref | GoogleScholarGoogle Scholar |
[27] Schooling, SR and Beveridge, TJ (2006) Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188, 5945–5957.
| Membrane vesicles: an overlooked component of the matrices of biofilms.Crossref | GoogleScholarGoogle Scholar |
[28] Schooling, SR et al. (2009) Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol 191, 4097–4102.
| Interactions of DNA with biofilm-derived membrane vesicles.Crossref | GoogleScholarGoogle Scholar |
[29] Johnston, EL et al. (2023) Planktonic and biofilm-derived Pseudomonas aeruginosa outer membrane vesicles facilitate horizontal gene transfer of plasmid DNA. Microbiol Spectr 11, e05179-22.
| Planktonic and biofilm-derived Pseudomonas aeruginosa outer membrane vesicles facilitate horizontal gene transfer of plasmid DNA.Crossref | GoogleScholarGoogle Scholar |
[30] Secor, PR et al. (2015) Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18, 549–559.
| Filamentous bacteriophage promote biofilm assembly and function.Crossref | GoogleScholarGoogle Scholar |


