Promoter targeted small RNAs: stabilising viral reservoirs
Anthony D KelleherSt Vincent’s Centre for Applied Medical Research, Sydney, NSW, Australia
Kirby Institute, UNSW, NSW, Australia
Tel: +61 2 8382 4941
Fax: +61 2 8382 4967
Email: t.kelleher@amr.org.au
Microbiology Australia 35(2) 103-104 https://doi.org/10.1071/MA14033
Published: 16 April 2014
The viral reservoir, a major barrier to curative therapy for HIV-1 infection, consists of viral DNA sequestered in long-lived resting CD4+ T cells and macrophages in diverse anatomical sites. Some of these sites are recognised parts of the immune system, such as lymph nodes. Others, such as the brain are not1. Current anti retroviral therapy (ART) does not substantially impact upon this reservoir. Early intervention with ART limits the reservoir but does not result in its elimination2–5.
This reservoir is believed to consist of integrated virus, which is in a latent state, silenced by epigenetic modifications of histone tails in the nucleosomes associated within the viral promoter, the 5’LTR. These histone tail modifications, which include deacetylation and methylation of lysines 9 and 27 of histone 3 (H3K9 and H3K27), are induced by the recruitment of enzymes such as histone deacetylases (HDACs) and histone methyltransferases6,7.
Most current efforts aimed at modulating the reservoir have attempted to reverse this silencing. However, trials of global activation of T cells by cytokine stimulation or T cell receptor stimulation have failed8,9. An alternate more recent approach has been to reverse latency via HDAC inhibitors such as valproate, vironostat and panabinostat10,11. These have induced modest increases in viral transcription, but this alone does not appear potent enough to substantially impact upon the reservoir. Further, efficacy of this approach is likely limited by non-specific gene activation and by the fact that viral latency is determined by complex epigenetic machinery of which HDACs are only one component12,13.
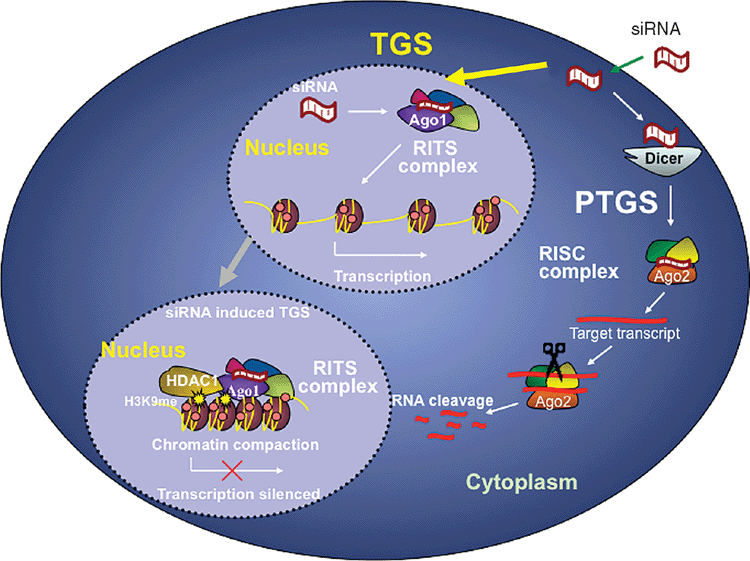
An alternate, far less explored, approach is to stabilise the reservoir by enforcing viral latency. This can be achieved by short interfering (si) or short hairpin (sh)RNAs targeting the 5’LTR of HIV-1. These short double stranded RNAs have the ability to induce transcriptional gene silencing (TGS) of the virus14–16 (Figure 1). The virus is unable to produce any mRNA transcripts from its genome and therefore cannot replicate. TGS, as distinct from siRNA induced post-transcriptional gene silencing of HIV-1, is long lasting and relatively resistant to escape. It produces far greater and more sustained decreases in viral load than the most promising PTGS inducing siRNAs14,17,18. A single dose of siRNAs targeting the highly conserved, unique sequences of the tandem repeat of the NF-κB binding motif, which is located within the non-transcribed portion of 5’LTR, completely silences HIV-1 replication in vitro for >30 days14. When delivered by retrovirus, as a shRNA, it silences the virus for >1 year in vitro18. The effect of these constructs is highly specific. They do not interfere with cell growth or viability nor do they change the expression of host genes regulated by the transcription factor NF-κB, nor do they induce an inflammatory response through induction of interferons19.
These constructs turn off viral transcription by inducing and then enforcing epigenetic changes similar to those seen in viral latency. They induce a closed chromatin structure within the HIV-1 genome associated with methylation of H3K9 and H3K27, and recruitment of HDACs, resulting in nucleosome rearrangement and chromatin compaction and loss of RNA polymerase II17,18,20. The RNA binding protein, Argonaute 1, complexed with the antisense strand of the siRNA is critical to this process, conferring sequence specificity and allowing preferential translocation to the nucleus of infected cells21.
Effective delivery of these molecules is perhaps the greatest challenge facing the clinical development of this approach. Recently, it has been demonstrated that these complexes can be delivered as short hairpin RNAs after lentiviral transduction of CD4 T cells, and suppress virus even in in vivo models of uncontrolled HIV infection22. Currently it is envisaged that, as with other gene therapies of this type, these si/shRNAs would be administered as cell therapy to patients who have sustained viral suppression on ART. Once the cells have engrafted, ART would be ceased and the shRNAs produced from these cells would maintain viral latency, enforcing prolonged viral latency and perhaps a functional cure.
However, the optimisation of this process is not trivial. Currently, like all cell therapies, this involves harvesting CD4+T cells and/or CD34+ stem cells from virally suppressed patients by leukapheresis, transduction of these cells with the gene therapy constructs in a purpose built laboratory, followed by reinfusion of these cells into patients. While this is challenging, the process is even more complicated. These cells need help to engraft effectively. Currently, the only available approach to this requires conditioning of the bone marrow with doses of chemotherapeutic drugs. This is a real challenge, even though the doses given are substantially reduced. Novel means of enhancing engraftment or alternative delivery vehicles such as nanoparticles or liposomes may simplify this process and are being explored. Therefore, although this approach is encouraging, like all the current strategies aimed at stabilising the reservoir, there is much work to do and several substantial hurdles to clear before it is proven and practical therapy.
References
[1] Kelleher, A.D. and Zaunders, J.J. (2006) Decimated or missing in action: CD4+ T cells as targets and effectors in the pathogenesis of primary HIV infection. Curr. HIV/AIDS Rep. 3, 5–12.| Decimated or missing in action: CD4+ T cells as targets and effectors in the pathogenesis of primary HIV infection.Crossref | GoogleScholarGoogle Scholar | 16522253PubMed |
[2] Murray, J.M. et al. (2014) HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J. Virol. 88, 3516–3526.
| HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy.Crossref | GoogleScholarGoogle Scholar | 24403590PubMed |
[3] Murray, J.M. et al. (2012) Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing transmission afterwards. AIDS 26, 543–550.
| Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing transmission afterwards.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xjs1Wqs7k%3D&md5=44de74043705c1ccd6a000c963eea56aCAS | 22410637PubMed |
[4] Koelsch, K.K. et al. (2011) Impact of treatment with raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir. AIDS 25, 2069–2078.
| Impact of treatment with raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlGltrjM&md5=e3cd80bd92b239aa3c1340705d15695eCAS | 21860347PubMed |
[5] The SPARTAC Trial Investigators (2013) Short-course antiretroviral therapy in primary HIV infection. N. Engl. J. Med. 368, 207–217.
| Short-course antiretroviral therapy in primary HIV infection.Crossref | GoogleScholarGoogle Scholar | 23323897PubMed |
[6] Van Lint, C. et al. (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15, 1112–1120.
| 1:CAS:528:DyaK28XhvFGqsbs%3D&md5=491708c672b516a3d2339ecb2af5e21cCAS | 8605881PubMed |
[7] du Chene, I. et al. (2007) Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26, 424–435.
| Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXotFKntA%3D%3D&md5=8b86451d33d0df179bf099f3874aca41CAS | 17245432PubMed |
[8] van Praag, R.M. et al. (2001) OKT3 and IL-2 treatment for purging of the latent HIV-1 reservoir in vivo results in selective long-lasting CD4+ T cell depletion. J. Clin. Immunol. 21, 218–226.
| OKT3 and IL-2 treatment for purging of the latent HIV-1 reservoir in vivo results in selective long-lasting CD4+ T cell depletion.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXkvVSgurs%3D&md5=d70375975aca1cad520145a6c6e9d70bCAS | 11403229PubMed |
[9] Levy, Y. et al. (2009) Enhanced T cell recovery in HIV-1-infected adults through IL-7 treatment. J. Clin. Invest. 119, 997–1007.
| 1:CAS:528:DC%2BD1MXktlSntLw%3D&md5=8ee3fada35b0e268b46ba72ba8a1bac9CAS | 19287090PubMed |
[10] Routy, J. et al. (2012) Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 13, 291–296.
| Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1WlurbK&md5=55bbcd875a792084582b0f5745ac66d4CAS | 22276680PubMed |
[11] Archin, N.M. et al. (2012) Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485.
| Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFWjtrfP&md5=3232ff25692c00851930a16274cebb9eCAS | 22837004PubMed |
[12] Beliakova-Bethell, N. et al. (2013) Suberoylanilide hydroxamic acid induces limited changes in the transcriptome of primary CD4(+) T cells. AIDS 27, 29–37.
| Suberoylanilide hydroxamic acid induces limited changes in the transcriptome of primary CD4(+) T cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVWhtr3O&md5=9e809190ef6fc9dc0414faa5fa89bbc3CAS | 23221426PubMed |
[13] Suzuki, K. and Kelleher, A.D. (2010) Lessons from viral latency in T cells: manipulating HIV‐1 transcription by siRNA. Future Medicine 4, 199–213.
| 1:CAS:528:DC%2BC3cXjtVOru70%3D&md5=3b2faec89da07aef31d26a3cd7d60c6eCAS |
[14] Suzuki, K. et al. (2005) Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing 1, 66–78.
| 1:CAS:528:DC%2BD2MXhtF2lurfN&md5=896a494364f1e68022985c3abb40d7ddCAS | 19771207PubMed |
[15] Morris, K.V. et al. (2005) Inhibition of HIV-1 replication by siRNA targeting conserved regions of gag/pol. RNA Biol. 2, 17–20.
| Inhibition of HIV-1 replication by siRNA targeting conserved regions of gag/pol.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFSrsLvP&md5=a0d41488cae3f89df41a0256aaf692baCAS | 17132935PubMed |
[16] Singh, A. et al. (2014) Long-term suppression of HIV-1C virus production in human peripheral blood mononuclear cells by LTR heterochromatization with a short double-stranded RNA. J. Antimicrob. Chemother. 69, 404–415.
| Long-term suppression of HIV-1C virus production in human peripheral blood mononuclear cells by LTR heterochromatization with a short double-stranded RNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXlvFOrsw%3D%3D&md5=12a10b3fe63ffd333901d728485c8d7cCAS | 24022068PubMed |
[17] Suzuki, K. et al. (2008) Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J. Biol. Chem. 283, 23353–23363.
| Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXpvFGgtrw%3D&md5=cfa31d74615cccd5def7822ba65a1102CAS | 18519571PubMed |
[18] Yamagishi, M. et al. (2009) Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect. 11, 500–508.
| Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXkslyrsr8%3D&md5=c1520430e911740c9a6bda7b83e3b610CAS | 19233310PubMed |
[19] Suzuki, K. et al. (2011) Transcriptional gene silencing of HIV-1 through promoter targeted RNA is highly specific. RNA Biol. 8, 1035–1046.
| Transcriptional gene silencing of HIV-1 through promoter targeted RNA is highly specific.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XkslCjt7o%3D&md5=f224061820bd7eed814427134c4504a1CAS | 21955498PubMed |
[20] Weinberg, M.S. et al. (2006) The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12, 256–262.
| The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlSjs74%3D&md5=f43c6a0408d09f8700c805326ef17c5aCAS | 16373483PubMed |
[21] Ahlenstiel, C.L. et al. (2012) Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 40, 1579–1595.
| Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjtlGktrY%3D&md5=43f25d44a886afc8f4d3e90279fee199CAS | 22064859PubMed |
[22] Suzuki, K. et al. (2013) Promoter targeting shRNA suppresses HIV-1 infection in vivo through transcriptional gene silencing. Molecular Therapy – Nucleic Acids 2, e137.
| Promoter targeting shRNA suppresses HIV-1 infection in vivo through transcriptional gene silencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXkvVKiu7k%3D&md5=bd362f70f47adaf5bd67a830683dccc7CAS | 24301868PubMed |
Biography
Anthony Kelleher is a clinician scientist whose interests include the dissection of the immunovirology of HIV-1 infection to inform the development of vaccines and novel therapies.