Robert Gerard (Gerry) Milton Wake (1933–2020)
Ronald J. Hill A , Richard I. Christopherson A and Philip W. Kuchel
A , Richard I. Christopherson A and Philip W. Kuchel  A *
A *
A
Abstract
Robert Gerard (Gerry) Wake was born at Wangi Wangi, a town on the shore of Lake Macquarie, New South Wales, on 8 August 1933. He died from the complications of Parkinson Disease in Sydney on 26 January 2020. Wake had a long and distinguished academic career in biochemistry and molecular biology at the University of Sydney, spanning 1950—with a two-year postdoctoral period and various study leaves—to 1999. His association began as an undergraduate, progressing to MSc (1956) and PhD (1958) and an appointment as lecturer in 1961, and a professorship in 1977. He is internationally renowned for work on the physical biochemistry of the transformation of the milk protein κ-casein by the enzyme rennin; and what was to become his major career focus, the molecular biology of bacterial DNA replication; having been the first to show that the chromosome in Bacillus subtilis (the hay bacterium) is circular. This was only the second bacterium for this general characteristic of bacterial genomes to be discovered. Furthermore, and against conventional wisdom, he demonstrated that replication of the B. subtilis circular chromosome is bidirectional involving two replication forks moving away from a common origin. Wake was a dedicated educator at the undergraduate level and led by example with excellent postgraduate guidance. He also made major contributions to academic governance at the University of Sydney, and more broadly to science in Australia, through influential roles in the Australian Society for Biochemistry and Molecular Biology, the Australian Research Council, and the Australian Academy of Science.
Keywords: bidirectional replication, biology, casein, circular chromosomes, DNA binding proteins, DNA replication, genetics, κ-casein, life sciences, molecular biology, PhD student supervision, research led education.
Family and educational background
Gerry Wake was born to Cora Augusta Wake (née Patterson) and Milton Wake. His father was a business manager, and farm administrator whose jobs took the family to several locations in New South Wales, including Tamworth, and Orange. Gerry was the second son of three boys and five girls.
Wake's secondary schooling, along with that of his older brother Peter, was at St Bernard’s College, a boarding school in Katoomba in the Blue Mountains, New South Wales. He finished high school in 1950 as dux of his class, and was awarded a scholarship to attend university.
Wake took up the scholarship at the University of Sydney studying for a degree in science. He was one year behind Peter who had enrolled in medicine. Wake learnt from Peter about a challenging new subject called biochemistry, which immediately piqued his curiosity. He proceeded to an honours degree and then MSc and PhD in the well-equipped CSIRO Biophysical Unit within the Biochemistry Department. The work was carried out under the fastidious direction of Dr Hugh McKenzie (1923–2008).1 Malcolm Smith, also in the unit at the time, introduced Wake to the operation of advanced biophysical instruments including the analytical ultracentrifuge, which was used to measure the molecular weights of proteins, and to investigate protein-protein interactions. Malcolm and Hugh became firm friends with Wake, who at one stage spent weekends helping Malcolm to build his house. The research experience included purification and physical characterisation of ovalbumin, serum albumin and casein proteins. Thus, Wake developed the discipline and rigour that is characteristic of the physical sciences, but applied them to the study of products of living systems; this typified the research approach he took throughout his academic career. He graduated MSc in 1956 and PhD in 1958.
Next phase: beginning of an academic odyssey
Wake married Aileen Mary Carmichael (mother, Catherine; father, Marmaduke) in 1957. Aileen, a music teacher, helped make this a creative family environment in which to raise two children and to span sixty years. Once Wake completed his PhD, the young couple moved to the University of Wisconsin, at Madison, Wisconsin, where Wake joined the group of the young physical biochemist Robert L. (Buzz) Baldwin (1927–2021).2 They analysed casein and casein fractions by the then new technique of starch-urea gel electrophoresis, to unravel the complexity of this system, thus bringing a considerable advance to the understanding of these proteins.3 In 1959, when Baldwin was lured by Arthur Kornberg to the embryonic Department of Biochemistry at Stanford University, California, Wake was invited to accompany him (Fig. 1). This was the year in which Kornberg was awarded the Nobel Prize in Physiology or Medicine for the isolation, only three years before, of DNA polymerase, the enzyme that faithfully copies the base sequence of DNA.
Gerry Wake (at right) with Buzz Baldwin, Stanford University, 1961. A Wake family photograph.

At Stanford, Wake and Baldwin also began research that was inspired by what has been termed by some ‘the most beautiful experiment in Biology’—that of Meselson and Stahl demonstrating the semi-conservative replication of DNA in bacterial cells, employing the incorporation of a heavy isotope to track newly replicated DNA by a change in density.4 Wake and Baldwin aimed to show that the DNA polymerase purified by Kornberg also replicated DNA semi-conservatively in a test tube. Their biophysical experiments successfully demonstrated this. However, differences between the results in the whole organism and in a test tube appeared. In order to reconcile these differences they used a dAT copolymer template, with 5-bromouracil to density label the replicated strand and then both density gradient analytical ultracentrifugation and DNA thermal melting experiments were performed.5 These analyses led to the correct conclusion of semi-conservative replication in vitro and to Wake's insights that were to guide his subsequent research into the mechanism of chromosome replication in Bacillus subtilis, back at the University of Sydney.
Return to Australia: κ-casein and stabilisation of milk micelles
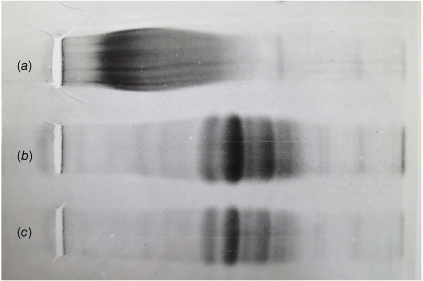
On their way to Sydney in 1961 the young couple spent six months in Oxford where Wake worked with Dr Rupert Cecil, an expert on the chemistry of sulfur-containing amino acids in proteins. This experience provided Wake with methods to make another critical advance in the casein field. Specifically, Wake and Baldwin had resolved casein fractions into species of different molecular weights by starch-urea gel electrophoresis; these fractions appeared as a sequence of discrete bands along with an enigmatic variable smear that represented what was the kappa (κ)-casein component. κ-casein was believed to correspond to a ‘stabilising colloid’ previously postulated by Linderstrøm-Lang to stabilise other caseins in the micelles that give milk its ‘milky’ appearance.6 This κ-casein component appeared to behave as a complex continuum of components that defied further conventional analysis. Based on his Oxford experience, Wake treated κ-casein with disulfide-bond cleaving reagents; the κ-casein smear dramatically resolved into discrete bands (Fig. 2).7 This led the way to a thorough understanding of the stabilisation of casein micelles and enzymatic processes involved in milk clotting and digestion in the gastrointestinal tracts of newborn and adult mammals.
Micelle-stabilising κ-casein presents an enigmatic continuum of molecular species on starch-urea gel electrophoresis (a). However, this protein resolves into a finite series of bands following reduction of disulfide bonds by mercaptoethanol and alkylation under denaturing conditions employing (b) 3 M guanidine hydrochloride, or (c) 8 M urea. Reproduced with the permission of R. J. Hill, MSc Thesis, University of Sydney.
New role: research-led education
Wake's first PhD student, Antony (Tony) Mackinlay, worked with him on casein aggregation and demonstrated convincingly that the ‘smear’ on electrophoretic analysis results from the crosslinking of discrete κ-casein monomers by disulfide bonds. Mackinlay went on to examine the action of rennin on the discrete components and focussed his project on the purification and characterisation of S-carboxymethyl derivatives of the newly recognised individual species, providing a basis for all subsequent studies of the κ-casein complex.8 Wake's second graduate student, Ronald (Ron) Hill, in the process of amino acid analysis of different regions of genetic variants of the κ-casein protein, noticed a biphasic distribution of hydrophobic and hydrophilic amino acid residues: the N-terminal region of the molecule was hydrophobic while near the C-terminus the hydrophilicity increased markedly with one residue in three of the last fifty eight being a hydroxylic amino acid, serine or threonine. This led Hill to postulate that the κ-casein polypeptide chain was functioning as an amphiphile to stabilise casein micelles with the hydrophobic N-terminal region binding to the hydrophobic core of the micelle, and the hydrophilic C-terminal region interacting with water, providing a hydrophilic shell. Rennin needs to split only one peptide bond in κ-casein, between the hydrophobic and hydrophilic regions, to destabilise the whole casein micelle system, thus leading to the separation of ‘curds and whey’! Hill and Wake showed (by using analytical ultracentrifugation) that the hydrophobic N-terminal fragment of κ-casein indeed interacts with a core casein component, but the hydrophilic C-terminal fragment does not. This was the first report of a protein functioning as an amphiphile, akin to the action of surfactant detergents. It was observed that the virus surface-located C-terminal region of the protein subunits of tobacco mosaic virus also exhibit a similar distribution of hydrophilic amino acids, suggesting a general principle underlying the stabilisation of macromolecular assemblies.9
Shift of research focus at Sydney: B. subtilis chromosome replication
Wake was now armed with insights from the work in Stanford with Baldwin, demonstrating the semiconservative replication of DNA molecules in vitro; and his experiments with Dale Kaiser and Ross Inman demonstrating concatemers of the bacteriophage λ chromosome, which are now considered obligatory intermediates in the replication of the virus.10 He was also inspired by the visualisation of individual replicating Escherichia coli chromosomes by John Cairns,11 leading him to shift his research focus to understanding the replication of bacterial chromosomes.
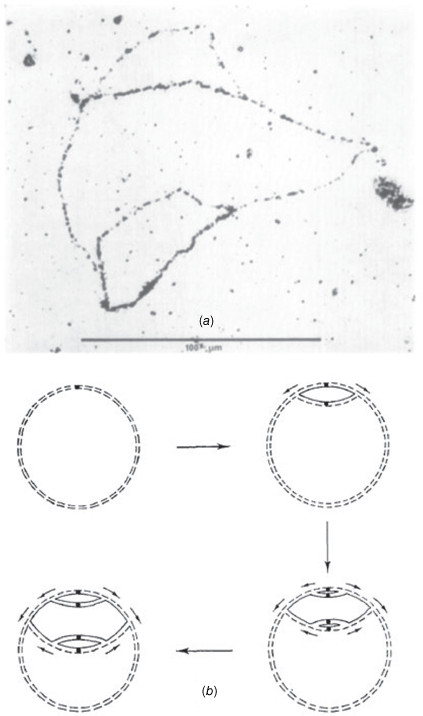
Wake had an uncanny ability to design experiments that enabled the elucidation of fundamental molecular mechanisms that underlie complex biological phenomena. A key example was his choice of the synchronously germinating B. subtilis spore system to study chromosome replication. He developed elegant experiments to monitor in parallel DNA synthesis and the transforming activity of genetic markers. His conclusion: ‘The synchrony in chromosome replication which accompanies emergence of the vegetative cell makes the germinating spore an attractive system for studying the events leading up to the initiation of DNA replication in bacteria’.12 This led to decades of highly successful fundamental research in his laboratory at the University of Sydney, encompassing demonstrating the circularity and bidirectional replication (Figs 3 and 4) and termination of replication of the B. subtilis chromosome (Fig. 5). This research area served as a training ground for many research students, and it attracted for sabbatical study Professor Jeff Errington FRS.13 Errington has recently been appointed as an ARC Australian Laureate Fellow at the University of Sydney.
(a) Wake's influential autoradiograph of symmetrically reinitiated B. subtilis chromosomes, demonstrating bidirectional DNA replication. Only the replicated DNA strands are radioactively labelled, allowing the replicated double-helical chromosomal regions to be visible in the autoradiograph as singly or doubly-labelled segments. (b) Scheme showing how the pattern of labelling in the autoradiograph arises from bidirectional replication. Original unlabelled polynucleotide strands are indicated by broken lines and newly replicated strands that have incorporated [3H]-thymine by unbroken lines. Reproduced from Wake (1972), Plate II (a); the article can be obtained free of charge via Open Access from Elsevier.
Gerry Wake with the microscope he used to demonstrate bidirectional replication of the B. subtilis chromosome by autoradiographic observation of individual replicating chromosomes, a discovery recognised in his citation for election to the Australian Academy of Science. Portrait by Judy Cassab (1999), property of the University of Sydney.

We leave it to the recollections of several of Wake’s former students to provide details of this research as recorded in the section headed ‘Pivotal personalities’ later in this memoir. For now, we step back and consider the more administrative aspects of Wake's academic life that enabled him to advance knowledge, and to teach it.
Emerging Biochemistry Department
Wake's 1953 BSc honours thesis was a 38-page typed document entitled ‘Report on fourth year research work submitted to the Department of Biochemistry: a study of the viscosity changes accompanying the urea denaturation of ovalbumin, serum albumin, chymotrypsin and ribonuclease’, supervised by Dr Hugh McKenzie. Wake's MSc (1956) and PhD (1958) projects were concerned with milk proteins and were also supervised by Hugh McKenzie; the work was the basis of the next phase of his career, forged by his overseas appointments.
After sojourns in Madison, Stanford and Oxford, Wake returned to the University of Sydney for his first academic appointment. Apparently, he had always aspired to return to Australia. The head of the Department of Biochemistry at the time was Professor John (Jack) L. Still; by 1960 he had been in this position for twelve years and was carefully building up an academic team. Therefore, it is relevant to detour into a brief history of the department to help underscore what a large span of its existence and achievements were associated with and influenced by Wake.
Department of Biochemistry: Wake’s roles
By 1938, Henry Priestley a Sydney medical graduate with overseas research experience had already served as acting professor of physiology after the retirement of the head of department in 1928. His research interests lay with vitamins and more generally with human nutrition; and from 1921 to 1928 he had been the McCaughey Associate Professor of Physiology (‘in the field of biochemistry’).14 Therefore, his appointment as professor of biochemistry in 1938 was a decisive step in the emergence of a separate discipline at the university. As McCaughey Professor of Biochemistry he was nevertheless still administratively embedded within the Department of Physiology but there was something that seemed to herald a long-term role for this discipline in university curricula, and it was the fact that Priestley’s chair was one of seven McCaughey Chairs that had been endowed by Sir Samuel McCaughey, a wealthy pastoralist, and were distributed across various faculties including arts, and now in perpetuity to biochemistry.
When Priestley retired in 1948, a separate administrative and academic Department of Biochemistry was born. Jack Still, who returned with a PhD from Cambridge (United Kingdom) was appointed to head the department/discipline. He set about building a group of staff to satisfy the burgeoning post-war hunger for the molecular understanding of living systems and therefore to teach these new ideas to science, engineering, medical, veterinary, pharmacy and dental students. As noted above, Wake entered this milieu in 1951. In the second year of his BSc studies, he first encountered Still who, as can be inferred from the above comments, had only been in his post for three years. Hence, Wake ‘grew up academically’ in the Department of Biochemistry almost from its inception as a stand-alone department.
Third McCaughey Professor of Biochemistry
With retirement looming and aiming to create a smooth transfer of responsibilities for the rapidly growing discipline, Still lobbied the senior administration of the university for a second ‘Establishment Chair’ of biochemistry. His argument was based on the escalating number of students who were keen to study this subject. The appointment of Professor Keith W. Taylor, an Englishman with active research interests in metabolism and diabetes, was made in 1975. Jack Still then retired (1976) and a search was made for a successor. Based on his emerging research excellence and all-round commitment to various academic posts, the selection committee looked no further than the in-house applicant, Gerry Wake. His appointment in 1977 was enthusiastically received by the University of Sydney community and the departmental members in particular. The rapidity of Wake's transition into one of the two top jobs in the department also made for a lot of efficiencies in administration. Keith Taylor was new to the University of Sydney system while Wake had a wealth of experience spanning his twenty years here. A nice touch for Wake was the transfer of the McCaughey Chair to him (because of his seniority and experience in the discipline), making him the third in the line of succession of this named chair.
Then, to the surprise of most, Taylor who was by now head of department and who had established a vibrant research group in the four years since his arrival, felt compelled to return to the United Kingdom for family reasons. This placed an extra teaching load on the staff and in particular on Wake, who then assumed the role of head of department. By 1980 after a two-year search the Chair Committee appointed Philip Kuchel who was then at the University of Newcastle, New South Wales. The employment contract specified that headship of the department would ‘rotate on a two year cycle … with Professor R. G. Wake’. Thus, armed with a formal agreement, the two professors alternated this responsibility until December 1997, when the department was merged with microbiology to form a new school. This period in the life of the Department of Biochemistry was characterised by hard work and harmony, in no small measure due to Wake's mantra expressed as follows: ‘If my research is going well, I can handle anything!’ Wake was an example to all who knew him for his lack of procrastination and thoughtful engagement in any committee of which he was a member.
Other aspects of the department: a cohort of research students
The role of Wake in the more general academic life of the department and his influence on others has been comprehensively recounted by Richard Christopherson and others.15
One of the privileges of academic life, that was cherished by Wake, is the moulding of ‘scientific character’ in aspiring students. He attracted a remarkable cohort of research students to his laboratory based on his engaged instruction and mentorship. Table 1 lists the Masters and PhD students he supervised over his career, all at the University of Sydney. The right-hand column indicates the career destinations of these graduates; almost all went on to leadership roles in science, with six of his PhD students and three Masters students ultimately becoming full professors. There had been a stream of academically outstanding BSc(Hons) students, some of whom proceeded on to PhD candidature with him. The broader group will not be dwelt upon here as their contributions to the steady development of science from the laboratory are covered both above and below, but a unique cohort of students who spent only one year with Gerry bears special mention. These were the five medical students who had signalled a deeper interest in the molecular aspects of medicine by taking a year out from their medical studies to obtain a BSc(Med). Table 1 includes these graduates each of whom rose rapidly to leadership positions in medicine in various roles. In recalling Wake they uniformly reflected on the profound influence that he had on their approach to biomedical research.
| Years | Name | Degree | Protein/DNA | Career destination | |
|---|---|---|---|---|---|
| 1962–3 | Antony Mackinlay | MSc | Protein | Associate Professor, University of New South Wales | |
| 1963–5 | PhD | ||||
| 1963 | Ronald Hill | BSc(Hons) | Protein | Chief Research Scientist CSIRO and Honorary Professor, University of Sydney | |
| 1964–5 | MSc | ||||
| 1965–68 | PhD | ||||
| 1964 | Liz Dennis AC | BSc(Hons) | DNA | Chief Research Scientist CSIRO, and CSIRO Fellow | |
| 1965–8 | PhD | ||||
| 1967 | Ruth Hall OAM | MSc | DNA | Professor, University of Sydney | |
| 1968 | Phillip Nagley AM | MSc | Initiation of replication | Professor, Monash University | |
| 1978 | Tony Weiss AM | BSc(Hons) | Termination of replication | McCaughey Professor of Biochemistry, University of Sydney | |
| 1979–84 | PhD | ||||
| 1982 | Tiina Iismaa | BSc(Hons) | Termination of replication | Career Scientist, Garvan Institute | |
| 1983–7 | PhD | ||||
| 1987–91 | Peter Lewis | PhD | Termination of replication | Professor, University of Newcastle NSW | |
| 1988 | Liz Harry | BSc(Hons) | Cell division | Professor University of Technology, Sydney | |
| 1989–1992 | PhD | ||||
| 1991–6 | Susan Rowland | PhD | Cell division | Professor and Vice-Provost, University of Sydney | |
| 1992 | David Langley | BSc(Hons) | Termination of replication | Career Scientist, Garvan Institute | |
| 1993–6 | PhD | ||||
| 1993–8 | Penny Young (now Andersen) | PhD | Termination of replication | Address unknown | |
| 1992–6 | Alison Franks | PhD | Termination of replication | Secondary science teacher | |
| 1995–9 | Angela Griffiths (now Owens) | PhD | Termination of replication | Senior Lecturer, University of Notre Dame | |
| 1995 | Vittorio Katis | BSc(Hons) PhD | Cell division | Career scientist, Oxford, UK | |
| 1996–9 | |||||
| 1996 | Iain Duggin | BSc(Hons) | Termination of replication | Associate Professor, University of Technology, Sydney | |
| 1997–2001 | PhD | ||||
| 1983 | Mark Smith | BSc(Hons) | Termination of replication | Continued as Gerry’s Research Assistant—see belowA | |
| 1984–96 | |||||
| 1976 | Leslie Burnett AM | BSc(Med) | Termination of replication | Professor, University of Sydney, & University of New South Wales | |
| 1980 | Iswar Hariharan | BSc(Med) | Termination of replication | Professor, UC Berkley, USA | |
| 1983 | John Rasko AO | BSc(Med) | Termination of replication | Professor, University of Sydney | |
| 1985 | Dominic Rowe | BSc(Med) | Termination of replication | Professor, Macquarie University | |
| 1990 | Roderick Clifton-Bligh | BSc(Med) | Termination of replication | Associate Professor, University of Sydney |
Pivotal personalities
While space restrictions preclude all the comments that we have received from graduates of Wake's laboratory, it is historically valuable to convey some of their thoughts. By doing this, the evolution of science that emerged from the laboratory is given a human/personal dimension, in chronological order of their respective candidatures:
Tony Mackinlay was Wake's first graduate research student. Mackinlay played an important role in setting up the laboratory and then demonstrating that the complex smear displayed by κ-casein on gel electrophoresis was able to be resolved into discrete bands following disulfide-bond cleavage.16 Mackinlay showed that all of the components are subject to attack by rennin. He then purified and characterised the individual components of κ-casein, demonstrating the contribution to micro-heterogeneity played by post-translational modifications involving addition of carbohydrates and sialic acid. Furthermore, it was shown that both modified and non-modified κ-casein components stabilise milk micelles.17 His project provided the basis for all subsequent work on the κ-casein protein complex.
Ronald Hill, Wake's second graduate student, further characterised the components of κ-casein, their genetic variants and rennin cleavage products.18 Working in the CSIRO Division of Animal Genetics to localise amino acid replacements in κ-casein genetic variants, Hill discovered an unusual composition with the C-terminal 58 amino acid residues presenting approximately one third as serine or threonine along with a similar proportion of aspartic and glutamic acids (and/or their amides); together these amino acids make up more than 50% of the total. In contrast, the N-terminal region of the molecule is predominantly hydrophobic. This distribution of residues suggested that κ-casein acts as an amphiphile to stabilise the hydrophobic core of casein micelles in an aqueous environment. This discovery was the first recognised example of a protein polypeptide chain acting as an amphiphile. It was reported in Nature,19 where it sets out a principle that also provides an explanation for the stability of other macromolecular assemblies involving protein-aqueous interfaces, including tobacco mosaic virus particles and the hydrophobins.20
The mindset and focus on molecular mechanism can be clearly seen in the context of this first major independent scientific foray. What followed was to become Wake's most famous body of work and it began with the honours project conducted by:
Elizabeth (Liz) Dennis (also known as Goldring) who wrote:
‘I came to Gerry’s lab to do honours in 1964, keen to be part of the molecular biology revolution. For my PhD, I initiated studies on the physical structure of the B. subtilis genetic material. It wasn’t clear how bacterial DNA was organised; viz, was all the DNA in a single chromosome and did it replicate from a single origin? I estimated the size of the B. subtilis genome by measuring the DNA content per spore.21 I approached the organisation of the genome using autoradiography of tritium-labelled DNA obtained from gently lysed cells, hoping to maintain the integrity of the DNA molecules. Structures containing up to 70% of the total DNA were observed indicating that at least this much of the genome was organised into one chromosome.22 As these structures did not appear to be intact chromosomes it was likely that they arose from breakage of a single chromosome comprising the entire genome. I also obtained evidence for the chromosome(s) being circular.23 Gerry was pleased with this work, as setting up the autoradiography provided a technique for further studies on the B. subtilis genome.24 Only the E. coli genome had been studied at this time, so this work enabled B. subtilis to feature as another model organism.’
This autoradiographic technique subsequently enabled Wake to demonstrate that replication of the B. subtilis chromosome was bidirectional, involving two replication forks proceeding in opposite directions from an origin, against conventional wisdom at the time (Fig. 3).25
Ruth Hall joined Gerry’s Laboratory in 1967 and completed an MSc before moving to the United Kingdom for a PhD. Following her own remarkable career in bacterial genetics, with the discovery of integrons (critical in enabling the spread of antibiotic resistance) she returned in 2003 to the molecular bioscience building that Jack Still had so effectively fought for when establishing biochemistry as a ‘stand-alone’ discipline at the University of Sydney. Hall's project work with Wake was, in part, contemporaneous with those of Dennis and Phillip Nagley and is described in general terms by them. Specifically:
Phillip (Phil) Nagley enrolled for and completed a notable MSc study in Wake's laboratory in 1968. His project arose from an observation made by Wake in the course of demonstrating extremely low, if any, DNA repair synthesis as opposed to replication in germinating B. subtilis spores.26 Wake separated un-replicated from replicated DNA by incorporating bromodeoxyuridine (BU) to increase the density of the latter in caesium salt gradients generated in analytical and preparative ultracentrifuges. Un-replicated DNA contained two ‘light’ strands (LL), once-replicated DNA contained one light and one heavy strand (LH), and twice replicated DNA contained two heavy strands (HH). Nagley recalled:
‘Gerry wondered if an HH peak that could be seen in caesium salt gradients after 150 min of spore germination was due to asynchronous initiation of the first and subsequent rounds of DNA replication (perhaps caused by the use of BU in the medium), or if the HH peak arose from a specifically timed initiation of a second round of DNA synthesis. My project was designed to analyse in detail the initiation of DNA replication in germinating spores, in the presence of BU.
‘I applied density gradient ultracentrifugation in the Beckman Model E ultracentrifuge to analyse the distribution of LL, LH and HH DNA in Cs2SO4 gradients after different times of germination in the presence of BU and thymine (T). I determined that the initiation of the first round of DNA replication took place at about 60 min and a second round was initiated at about 130 min after germination. These two rounds proceed in parallel about 70 min ‘out of phase’ from one another. I demonstrated that the second round took place at the same initiation point as that of the first and proceeded in the same direction, by using BU and T in different ratios in the medium, adjusted at various times after germination. I calculated that the standard BU-T medium I was using slowed down the DNA replication as a whole by about 40% and calculated the normal time for complete DNA replication in T medium (without BU) to be 62 min.
‘The critical experiments I carried out were to vary the rate of replication of the DNA as a whole by manipulating the total added BU + T in the medium, such that T and its analogue BU at lower concentrations became rate-limiting for DNA synthesis. Despite DNA replication being slowed by a further forty percent over the standard BU-T medium, the timing of initiation of neither the first nor the second round of replication was appreciably affected. I was able to confirm that in T medium alone (without BU), using transforming marker ratio analysis (for genetic markers near the chromosome origin, the middle of the chromosome and near the terminus), that the first round of DNA replication initiated about 60–70 min after germination and the second round some 60–70 min later (about 120–130 min after germination). This implied that in non-limiting amounts of T alone the initiation of the second round did not take place until replication of the first round was finished (itself taking about 62 min). But in conditions where BU was included and replication slowed, so-called ‘dichotomous’ replication occurred where a second set of forks were symmetrically opened up as DNA replication events, even before the first round had been completed.’27
Wake liked the conclusion that the second round of initiation was essentially normal and able to be observed in his system (apparently prematurely) purely due to the slowed rate of replication in the presence of BU. These findings vindicated his use of the germinating spore system in B. subtilis to enable orderly studies on the initiation of DNA replication in bacteria thereby underpinning his later creative work on this system including that leading to the establishment of chromosome circularity and bidirectional replication (Figs 3 and 4).28
The use of sporulation in B. subtilis as a synchronising mechanism for studies of DNA replication led to the next major advance that was delivered by:
Anthony (Tony) Weiss who wrote:
‘My first experience in Gerry’s lab was as his Honours student in 1978 when I explored the progression of bidirectional replication forks as they approached the termination of chromosome replication. This work overlapped with Gerry’s sabbatical visit to Oxford to work with Professor Joel Mandelstam. Very little was known of how replication forks behave as they collide, and it was inevitable that Gerry’s elegant B. subtilis spore germination approach would be used. I transitioned to a PhD that commenced in 1979, and in a series of studies, we used synchronous replication in germinating spores to radiolabel the terminus region. We were surprised and delighted to find that termination was confined to a narrow part of the chromosome, which we named TerC. Our first publication co-authored with Iswar Hariharan was in Nature.29 Benefitting from Gerry’s attentive supervision, I generated a restriction map of TerC30 that was then used to discover that bidirectional DNA replication was sequential: one fork reached TerC first where it waited for the arrival of the second fork. Then Mark Smith and Tiina Iismaa helped clone DNA from this region.31 The finding of a unique DNA intermediate that is associated with termination of replication was published in Cell;32 this structure we suspected was a stalled replication fork.33
‘I remember vividly the effort needed to isolate enough of the putative stalled fork from countless agarose gels, which were then sent to Gerry’s friend Ross Inman in Wisconsin, who trawled through vast numbers of electron microscope images of DNA strands. To the team’s delight, Inman saw and measured the dimensions of the forks; they matched our predicted model-size precisely. This led to a collaborative publication in the Journal of Molecular Biology34 which transformed the field of the regulation of termination of bacterial DNA replication.’
Amongst other aspects of this work, Wake was especially impressed with Weiss's visual detection of a palindrome in the termination region of the chromosome. Much was now understood about the topology of the termination region, and this led to the next phase of the work.
Tiina Iismaa joined the now very active research group in 1983 to study for her BSc(Hons), for which she gained the University Medal, and proceeded to a PhD project in 1984. She wrote:
‘The restriction endonuclease map of the terminus region of the B. subtilis chromosome was confirmed and extended to a length representing approximately 4% of the chromosome.35 Genetic orientation of the restriction map allowed definitive identification of the clockwise-moving replication fork as the first to encounter the terminus, TerC, and to be arrested or impeded in its movement at this site. Three strains of B. subtilis that carry large deletions of the terminus region, including deletion of the replication fork arrest site TerC were demonstrated to exhibit normal growth rate and morphology when grown in complex medium, indicating the dispensability of TerC for vegetative growth and cell division.36 The construction and physical and physiological characterisation of a strain of B. subtilis that carries a smaller, defined deletion of TerC (strain SU153), confirmed dispensability of TerC for vegetative growth, cell division and for the specialised partitioning of the sporulating cell that is associated with the formation of the B. subtilis endospore. Reinsertion into the chromosome of strain SU153 of a 1.75 kb fragment of DNA spanning TerC, at a site approximately 25 kb closer to the approaching clockwise-moving replication fork and in the same orientation as in the wild-type chromosome, demonstrated that there had been no alteration with respect to TerC function in the cloned DNA that contains TerC, and that the 1.75 kb sequence, isolated within new flanking sequences, was sufficient to achieve replication fork arrest.’37
After Ismaa’s work, the mantle for the next stage in the evolution of understanding the termination of replication of the bacterial genome was passed to a PhD student who arrived from the United Kingdom.
Peter Lewis wrote:
‘My project continued the work by Tony Weiss and Tiina Iismaa on the mechanism of termination of chromosome replication in B. subtilis. This involved establishing that termination systems were present in other bacilli,38 purifying the replication terminator protein (RTP), and establishing its DNA binding site, and kinetics.39 By the time I undertook my project Gerry had been interested in termination of replication for many years, and a result that particularly pleased him was my first demonstration of the DNA-binding activity of RTP to the inverted repeat region (IRR) at the terminus (TerC). Previously this IRR had been identified visually by Tony Weiss. The first gel not only established the binding specificity of RTP, but it also demonstrated that there were four RTP binding sites, and cooperativity occurred between the RTP binding sites on the IRR. In the final part of my PhD work I showed that RTP functioned in a polar manner to inhibit the progress of DNA helicase at the apex of the replication fork; this enabled the formulation of a model for polar replication fork arrest.’
The results obtained by Lewis laid the foundation for the projects of all subsequent students including David Langley, Penny Anderson, Alison Franks, and Iain Duggin.
By the mid-1980s, Wake had developed an interest in the molecular basis of the coordination of DNA replication in B. subtilis with the binary fission (division) of the cell. The first student charged with exploring this area was:
Elizabeth (Liz) Harry who wrote:
‘In my Honours year (1986) we were the first to identify genes involved in cell division in B. subtilis, which we named ftsZ and divIB. FtsZ is now known to be the bacterial precursor of eukaryotic tubulin.
‘For my PhD work, we characterised DivIB, showing that it is a membrane protein, oriented towards the cell wall.40 We identified functional regions of the protein and showed that it had homologues in other bacteria; this latter finding took a while to achieve as bioinformatics was just emerging at the time! As a postdoc in Gerry’s lab in the mid 1990s, we used new cell biology techniques for bacteria, that were developed in Harvard during my postdoc there, to show that DivIB and several other division proteins localise to the division site; and we established their localisation dependency and order of assembly to create the divisome, as it is now called.41 Using these bacterial imaging techniques, Gerry, the team, and I focused attention on the regulation of cell division and its coordination with DNA replication. This led to the discovery that the initiation stage of replication is linked to the first stage of cell division, the assembly of the Z-ring (polymerisation of the FtsZ protein) at mid-cell.42 We also showed that positioning of the division site in B. subtilis does not depend on the Min system of proteins—overturning a long-held belief in the field.43
‘We often used the spore germination system in our experiments as it provided a single synchronous cell cycle, making it possible to identify how cell cycle processes are coordinated. Gerry was a specialist in this technique and his ability as a scientist to really get to the core of things, to identify the approach that would enable a clear-cut answer to the question, is what he excelled at.’
The work on termination of replication progressed rapidly, as well as that on cell division, and international attention was drawn to RTP. As sometimes happens, a new student added fresh impetus…
David Langley (Honours 1992, PhD 1993–6) wrote:
‘By this stage the project was also attracting more international attention, and the crystal structure of RTP was solved by an American group.44 Then the crystal structure of RTP bound to one of its DNA sequence elements was solved by an Australian collaborator, Matthew Wilce and his group.45 The turn of the century roughly coincided with Gerry gradually winding down actively working in the termination of replication field. Gerry’s curiosity and appetite for discovery however was ever present and over the course of this main body of work his focus was gradually shifting back towards another of his long-term scientific passions—the genetics, challenges and mechanisms concerning cell division.’
Langley and Wake moved the field forward comprehensively. Langley wrote:
‘I built upon the landmark confirmation by Peter Lewis that the RTP protein product of a gene identified at the terminus of replication did in fact bind the nearby DNA sequence elements, just as predicted and that both the protein and DNA sequences were required to halt progression of a replication fork. What wasn’t understood were the finer details of how? While Peter had used the “DNaseI foot-printing” technique (a DNA protection experiment) to identify a block of DNA sequence to which RTP bound, the aim in my project was to use a much smaller probe or cutting agent—not the enzyme DNaseI but something the size of a water molecule, the hydroxyl radical. Hydroxyl radicals were used to treat radiolabelled DNA strands believed to contain sequences to which RTP bound. Hydroxyl radicals act upon a DNA strand by randomly removing a nitrogen base or nucleoside. Treatment was performed such that each labelled strand had a single gap, or single rung of the DNA ladder missing. RTP was then added to the gapped duplex and those strands that were rendered incapable of binding RTP due to the gap position were not ‘band-shifted’ when the sample was electrophoresed on a native gel. These unbound bands were excised and run on a sequencing gel next to a sequencing ladder to reveal the relative intensity of gap positions that interfered with protein binding. The result was a fine detail footprint of the relative importance of each DNA position to the binding interaction with RTP.46 The experiment inadvertently reported not only the relative importance of each base position within the DNA, but also revealed the order by which the nested ‘half sites’ within the DNA sequence elements were bound by subsequent molecules of RTP. This revealed that each termination sequence element comprised two halves, the one bound very tightly by RTP and the other bound less tightly. This was important from the perspective of understanding that approaching replication forks might encounter the tightly-bound RTP when approaching from one direction, thus becoming halted, or might disrupt the complex when approaching from the less-well bound orientation, thus explaining the observation that the replication fork blocks appeared to be polar in action.’
By the mid 1990s, Wake and his team had developed a detailed understanding of the main molecular players in the termination of chromosome replication. Now was the time to ask and answer some deeper, more fundamental questions that have a bearing on the control and coordination of DNA replication in all biological species. This was explored by Honours student (1996) and then Wake’s last PhD student:
Iain Duggin who stated:
‘Gerry was especially interested in determining how the action of RTP was polar—the RTP complex bound to DNA was only able to block the movement of the replication fork from one direction of approach.47 In 1996, an Editor of Nature invited Gerry to provide a ‘News and Views’ commentary on details of molecular mechanisms of replication termination in E. coli and B. subtilis, which he aptly entitled ‘Tussle with a terminator’.48 He had noticed that the recognition DNA sequences for RTP (called terminator or Ter sites) were asymmetrical in their base sequence and must have the information required to generate the polarity, because RTP and the DNA backbone are symmetrical dimers and by themselves could not generate the asymmetry required to explain the polarity of action. His group discovered that each Ter site formed a cooperative complex with two dimers of RTP—one strong binding site and one weak.49 Interestingly, variants of the Ter sequence that were more symmetrical (binding strongly to two dimers of RTP) could function in a bidirectional manner to block replication fork movement.50 Gerry’s work with Glenn King, Greg Ralston, Jackie and Matthew Wilce and others resulted in structures of parts of the RTP-DNA complex, eventually published in Nature Structural Biology,51 and models arose for how the polarity could be generated from the asymmetry of the DNA sequence.’52
Duggin’s contributions are summarised in his own words:
‘I focussed on the symmetry (and asymmetry) within the Ter sites and then aimed to address how this generated functional asymmetry in the whole complex, that formed in polar fork arrest. Was it the affinity difference between the two half-sites (plus the required cooperative affinity between them) that allowed polar fork arrest, or was it more related to the structural conformational differences of RTP from each side that sterically “jammed” or specifically inhibited the replication machinery (replisome)? We found that the RTP-DNA complex was exceptionally stable, with highly cooperative binding that stabilised the complex.53 By systematically substituting different mutant DNA half-sites, a correlation appeared between the affinity of binding over the half-site facing the arrested fork and the efficiency of fork arrest.54 However, it appeared that the high binding affinity was not the only factor controlling the efficiency of replication fork arrest, because we found that: (a) RTP-Ter did not function efficiently when placed into E. coli,55 suggesting that specific features of the replisome were involved; (b) some mutant Ter sites or RTPs with altered binding affinity or stability deviated significantly from the correlation;56 and (c) specifically-designed RTP fusion proteins bound normally to DNA but they were inefficient in fork arrest.57 Our results indicated that both the differential binding affinity and the conformation of RTP were important factors in determining its ability to arrest replication fork movement in a polar manner.’
After Wake’s retirement, he co-authored an influential review on how termination of DNA replication occurs in bacteria via the ‘replication fork trap,’ which he was so instrumental in defining (Fig. 5).58 This was Wake’s last major review of what was truly ‘A life’s major work’. The review settled various debates about interpretation of experimental results and made key predictions about the biological roles of RTP-Ter complexes and their counterparts in E. coli (and other bacterial) chromosome replications. Predictably, the review caused a flurry of highly visible new research that confirmed the group’s observations and predictions; it is a fitting testament to Wake in the field.
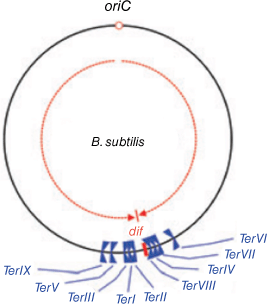
The circular B. subtilis chromosome depicting the directions of progress of the clockwise and anticlockwise replication forks, from the origin (in red) and the fork trap region comprising a series of Ter sites. TerI, TerIII, TerV and TerIX are oriented to impede clockwise moving forks, and TerII, TerVIII, TerIV, TerVII and TerVI are oriented to impede anticlockwise forks. Modified from data shown in Fig. 1B of Duggin and others (2008) entitled ‘The replication fork trap and termination of chromosome replication’. Reproduction of Fig. 1B from Duggin and others (2008) has been approved by John Wiley & Sons.
Social-scientific recognition and responsibilities
As might have been expected, with his scientific achievements becoming widely known in the field of bacterial molecular biology, Wake's work also became recognised more broadly and deserving of accolades. These flowed in Australia with invited lectures, prizes and awards.
He was awarded the Lemberg Medal of the Australian Biochemical Society in 1981, and served as president of the society from 1984 to 1986.
He was elected a Fellow of Australian Academy of Science in 1985, and he gave his time generously to the academy. He served on its council (1990–3) and as vice-president (1991–2). Wake served on the academy’s National Committee for Biochemistry as a member (1979–81) and then as chair (1982–5). He also served as a member of the Sectional Committee then responsible for Biology at the Cellular and Molecular Level (1986–9), as chair (1989–90), and as a member of the Sectional Committee then responsible for Biochemistry, Molecular Biology and Immunology (1996–2000). He also served as a member of the Boden Research Conferences selection committee (1996–9). He was chairman of the Scientific Programme Committee of the 7th Congress of the Federation of Asian and Oceanian Biochemists and Molecular Biologists, held in Sydney in 1995.
Wake was keen to help other scientists with the design of, and expressing the worth of, their research projects. He joined the Australian Research Grants (ARGC) Committee for Molecular Biology and served for two years, one with Jim Peacock who was responsible for running the committee. Peacock found that Wake gave valuable advice to all the research groups he interviewed, and in doing this job he set new ways for all the ARGC committees to operate with individual scientists and their research groups.
Legacies
It could be argued that Wake's greatest academic legacy lives on through the graduate students he trained in the ‘art of scientific research’. These students perhaps had some very general notions about the nature of research when they arrived in his laboratory. Wake taught by example that behind his elegant research there lay the power of logical deduction to understand nature’s organisation, planning and an uncanny ability to inspect complex phenomena to see underlying simpler mechanisms that enabled him (and his students) to focus their investigations on ‘key questions’. The approaches he imparted led his students onto highly successful careers in scientific research and education, many at professorial level, and including by way of example Fellowships of the Australian Academy of Science, Foreign Member of the United States National Academy of Sciences, Research Institute Directorships, Fulbright Scholarships, the Prime Minister’s Prize for Science, and the Prime Minister’s Prize for Innovation, the Clunies Ross Knowledge Commercialisation Award in Biotechnology, and recognition in the Order of Australia honours awards. The value of Wake’s conviction regarding ‘the value of basic research’ is amply illustrated not only by the senior academic careers of his graduate students but also by the major commercial manifestations flowing from their research.
After his retirement in 2000, Wake continued to play a role in academic life and to follow his more general interests including the interface between science and theology, woodworking, and tennis. However, he relished the additional time he could spend with his family. He passed away on 26 January 2020 after courageously facing the relentless decline associated with Parkinson Disease. He was a very fine person: a loving husband to Aileen (Fig. 6), a devoted father to Rachel and Simon; father-in-law to Essie; and grandfather to Oliver, Archie, Imogen, Juno and Finn. The large gathering of scientists, relatives, and friends, including those from his church, at Wake's funeral also attested to his inspirational legacy as father and grandfather in a large social community.
He will be greatly missed, while his human and scientific legacies live on. Wake’s full list of publications, in chronological order, is given in the Supplementary material.
Data availability
All data used in the compilation of this biographical memoir are publicly available and none was created specifically for the memoir.
Acknowledgements
We thank Gerry Wake’s former postdoctoral fellows and students, including especially those mentioned in the Pivotal Personalities section above for their first-hand insights; and to Susan Rowland and Glenn King for so thoroughly proof-reading the manuscript. Thanks also go to Ruth Hall who through circumstances beyond her control was compelled to pass authorship of this article to we three. Special thanks go to Aileen and Gerry’s family for access to archival material and family anecdotes, all fascinating and yet not all included here because of space restrictions.
References
Andersen, P. A., Griffiths, A. A., Duggin, I. G., and Wake, R. G. (2000) Functional specificity of the replication fork-arrest complexes of Bacillus subtilis and Escherichia coli: significant specificity for Tus-Ter functioning in E. coli, Molecular Microbiology, 36(6), 1327-1335.
| Crossref | Google Scholar | PubMed |
Armstrong, C. E., Mackinlay, A. G., Hill, R. J., and Wake, R. G. (1967) The action of rennin on κ-casein: the heterogeneity and origin of the soluble product, Biochimica Et Biophysica Acta, 140(1), 123-131.
| Crossref | Google Scholar |
Bussiere, D. E., Bastia, D., and White, S. W. (1995) Crystal structure of the replication terminator protein from B. subtilis at 2.6 Å, Cell, 80, 651-660.
| Crossref | Google Scholar | PubMed |
Cairns, J., and Sinsheimer, R. L. (1963) Bacterial chromosome and its manner of replication as seen by autoradiography, Journal of Molecular Biology, 6(3), 208-213.
| Crossref | Google Scholar | PubMed |
Christopherson, R. I., Hill, R. J., Weiss, A. S., and Kuchel, P. W. (2020) In memorium: Robert Gerard (Gerry) Wake FAA 1933–2020, Australian Biochemist, 51, 45-47.
| Google Scholar |
Dennis, E. S., and Wake, R. G. (1966) Autoradiography of the Bacillus subtilis chromosome, Journal of Molecular Biology, 15(2), 435-439.
| Crossref | Google Scholar | PubMed |
Duggin, I. G. (2006) DNA replication fork arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding, Journal of Molecular Biology, 361, 1-6.
| Crossref | Google Scholar | PubMed |
Duggin, I. G., Andersen, P. A., Smith, M. T., Wilce, J. A., King, G. F., and Wake, R. G. (1999) Site-directed mutants of RTP of Bacillus subtilis and the mechanism of replication fork arrest, Journal of Molecular Biology, 286(5), 1325-1335.
| Crossref | Google Scholar | PubMed |
Duggin, I. G., Matthews, J. M., Dixon, N. E., Wake, R. G., and Mackay, J. P. (2005) A complex mechanism determines polarity of DNA replication fork arrest by the replication terminator complex of Bacillus subtilis, Journal of Biological Chemistry, 280(13), 13105-13113.
| Crossref | Google Scholar | PubMed |
Duggin, I. G., Wake, R. G., Bell, S. D., and Hill, T. M. (2008) The replication fork trap and termination of chromosome replication, Molecular Microbiology, 70(6), 1323-1333.
| Crossref | Google Scholar | PubMed |
Goldman, B. (2021) Biochemist Robert Baldwin dies at 93, Stanford Medicine News, https://med.stanford.edu/news/all-news/2021/03/Robert-Baldwin-biochemistry-department-dies-at-93.html
Goldring, E. S., and Wake, R. G. (1968) A comparison of the segregation of chromosomes within microcolonies developing from single Bacillus subtilis and Bacillus megaterium spores, Journal of Molecular Biology, 35(3), 647-650.
| Crossref | Google Scholar | PubMed |
Harry, E. J., and Wake, R. G. (1989) Cloning and expression of a Bacillus subtilis division initiation gene for which a homolog has not been identified in another organism, Journal of Bacteriology, 171(12), 6835-6839.
| Crossref | Google Scholar |
Harry, E. J., Stewart, B. J., and Wake, R. G. (1993) Characterization of mutations in DivIB of Bacillus subtilis and cellular-localization of the DivIB protein, Molecular Microbiology, 7(4), 611-621.
| Crossref | Google Scholar | PubMed |
Harry, E. J., Rodwell, J., and Wake, R. G. (1999) Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly, Molecular Microbiology, 33(1), 33-40.
| Crossref | Google Scholar | PubMed |
Hill, R. J., and Wake, R. G. (1969) Amphiphile nature of κ-casein as basis for its micelle stabilizing property, Nature, 221, 635-639 221.
| Crossref | Google Scholar | PubMed |
Hill, R. J., Naughton, M. A., and Wake, R. G. (1970) The major genetic variant macropeptides of κ-casein—a comparisonof their amino acid contents and tryptic peptides, Biochimica Et Biophysica Acta, 200, 267-274.
| Google Scholar | PubMed |
Iismaa, T. P., and Wake, R. G. (1987) The normal replication terminus of the Bacillus subtilis chromosome, terC is dispensable for vegetative growth and sporulation, Journal of Molecular Biology, 195(2), 299-310.
| Crossref | Google Scholar | PubMed |
Iismaa, T. P., Smith, M. T., and Wake, R. G. (1984) Physical map of the Bacillus subtilis replication terminus region – its confirmation, extension and genetic orientation, Gene, 32(1–2), 171-180.
| Crossref | Google Scholar | PubMed |
Iismaa, T. P., Carrigan, C. M., and Wake, R. G. (1988) Relocation of the replication terminus, terC, of Bacillus subtilis to a new chromosomal site, Gene, 67(2), 183-191.
| Crossref | Google Scholar | PubMed |
Jeffrey, P., Kuchel, P., McKenzie, R. and Harris, K. (2024) Dr Hugh A McKenzie CChem FRSC, Royal Soc Chem, https://www.rsc.org/membership-and-community/member-obituaries/2005-2009/dr-hugh-a-mckenzie-cchem-frsc/
Kralicek, A. V., Wilson, P. K., Ralston, G. B., Wake, R. G., and King, G. F. (1997) Reorganization of terminator DNA upon binding replication terminator protein: implications for the functional replication fork arrest complex, Nucleic Acids Research, 25(3), 590-596.
| Crossref | Google Scholar | PubMed |
Langley, D. B., Smith, M. T., Lewis, P. J., and Wake, R. G. (1993) Protein-nucleoside contacts in the interaction between the replication terminator protein of Bacillus subtilis and the DNA terminator, Molecular Microbiology, 10(4), 771-779.
| Crossref | Google Scholar | PubMed |
Lewis, P. J., and Wake, R. G. (1989) DNA and protein-sequence conservation at the replication terminus in Bacillus subtilis 168 and Bacillus subtilis W23, Journal of Bacteriology, 171(3), 1402-1408.
| Crossref | Google Scholar | PubMed |
Lewis, P. J., and Wake, R. G. (1991) Termination of chromosome-replication in Bacillus subtilis, Research in Microbiology, 142(7–8), 893-900.
| Crossref | Google Scholar | PubMed |
Lewis, P. J., Smith, M. T., and Wake, R. G. (1989) A protein involved in termination of chromosome replication in Bacillus subtilis binds specifically to the terC site, Journal of Bacteriology, 171(6), 3564-3567.
| Crossref | Google Scholar | PubMed |
Lewis, P. J., Ralston, G. B., Christopherson, R. I., and Wake, R. G. (1990) Identification of the replication terminator protein-binding sites in the terminus region of the Bacillus subtilis chromosome and stoichiometry of the binding, Journal of Molecular Biology, 214(1), 73-84.
| Crossref | Google Scholar | PubMed |
Linder, M. B., Szilvay, G. R., Nakari-Setälä, T., and Penttilä, M. E. (2005) Hydrophobins: the protein-amphiphiles of filamentous fungi, FEMS Microbiology Reviews, 29(5), 877-896.
| Crossref | Google Scholar | PubMed |
Linderstrom-Lang, K. (1929) Studies on casein III. On the fractionation of casein, Comptes Rendus des Travaux du Laboratoire Carlsberg, 17(9), 1-116.
| Google Scholar |
Mackinlay, A. G., and Wake, R. G. (1964) Heterogeneity of κ-casein, Biochimica Et Biophysica Acta, 93(2), 378-386.
| Crossref | Google Scholar | PubMed |
Mackinlay, A. G., and Wake, R. G. (1965) Fractionation of S-carboxymethyl-κ-casein and characterization of the components, Biochimica Et Biophysica Acta, 104(1), 167-180.
| Crossref | Google Scholar | PubMed |
Meijer, W. J., Smith, M., Wake, R. G., deBoer, A. L., Venema, G., and Bron, S. (1996) Identification and characterization of a novel type of replication terminator with bidirectional activity on the Bacillus subtilis theta plasmid pLS20, Molecular Microbiology, 19(6), 1295-1306.
| Crossref | Google Scholar | PubMed |
Meselson, M., and Stahl, F. W. (1958) The replication of DNA in Escherichia coli, Proceedings of the National Academy of Sciences of the United States of America, 44(7), 671-682.
| Crossref | Google Scholar | PubMed |
Migocki, M. D., Freeman, M. K., Wake, R. G., and Harry, E. J. (2002) The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis, EMBO Reports, 3(12), 1163-1167.
| Crossref | Google Scholar | PubMed |
Nagley, P., and Wake, R. G. (1969) Effect of 5-bromouracil on the pattern of deoxyribonucleic acid replication in germinating Bacillus subtilis spores, Journal of Molecular Biology, 43(3), 619-630.
| Crossref | Google Scholar | PubMed |
Smith, M. T., and Wake, R. G. (1992) Definition and polarity of action of DNA-replication terminators in Bacillus subtilis, Journal of Molecular Biology, 227(3), 648-657.
| Crossref | Google Scholar | PubMed |
Wake, R. G. (1963) Sequential replication of DNA in synchronously germinating Bacillus subtilis spores, Biochemical and Biophysical Research Communications, 13(1), 67-70.
| Crossref | Google Scholar | PubMed |
Wake, R. G. (1967) A study of the possible extent of synthesis of repair DNA during germination of Bacillus subtilis spores, Journal of Molecular Biology, 25(2), 217-234.
| Crossref | Google Scholar | PubMed |
Wake, R. G. (1972) Visualization of reinitiated chromosomes in Bacillus subtilis, Journal of Molecular Biology, 68(3), 501-509.
| Crossref | Google Scholar | PubMed |
Wake, R. G. (1973) Circularity of the Bacillus subtilis chromosome and further studies on its bidirectional replication, Journal of Molecular Biology, 77(4), 569-575.
| Crossref | Google Scholar | PubMed |
Wake, R. G. (1996) DNA replication – tussle with a terminator, Nature, 383(6601), 582-583.
| Crossref | Google Scholar | PubMed |
Wake, R. G., and Baldwin, R. L. (1960) Gel electrophoresis and its application to analysis of the caseins, Food Technology, 14(6), B36.
| Google Scholar |
Wake, R. G., and Baldwin, R. L. (1962) Physical studies on the replication of DNA in vitro, Journal of Molecular Biology, 5(2), 201-216.
| Crossref | Google Scholar | PubMed |
Wake, R. G., and Errington, J. (1995) Chromosome partitioning in bacteria, Annual Review of Genetics, 29, 41-67.
| Crossref | Google Scholar | PubMed |
Wake, R. G., Kaiser, A. D., and Inman, R. B. (1972) Isolation and structure of phage λ head-mutant DNA, Journal of Molecular Biology, 64(3), 519-540.
| Crossref | Google Scholar | PubMed |
Weiss, A. S., and Wake, R. G. (1983) Restriction map of DNA spanning the replication terminus of the Bacillus subtilis chromosome, Journal of Molecular Biology, 171(2), 119-137.
| Crossref | Google Scholar | PubMed |
Weiss, A. S., and Wake, R. G. (1984a) Impediment to replication fork movement in the terminus region of the Bacillus subtilis chromosome, Journal of Molecular Biology, 179(4), 745-750.
| Crossref | Google Scholar |
Weiss, A. S., and Wake, R. G. (1984b) A unique DNA intermediate associated with termination of chromosome-replication in Bacillus subtilis, Cell, 39(3), 683-689.
| Crossref | Google Scholar |
Weiss, A. S., Hariharan, I. K., and Wake, R. G. (1981) Analysis of the terminus region of the Bacillus subtilis chromosome, Nature, 293(5834), 673-675.
| Crossref | Google Scholar | PubMed |
Weiss, A. S., Smith, M. T., Iismaa, T. P., and Wake, R. G. (1983) Cloning DNA from the replication terminus region of the Bacillus subtilis chromosome, Gene, 24(1), 83-91.
| Crossref | Google Scholar | PubMed |
Weiss, A. S., Wake, R. G., and Inman, R. B. (1986) An immobilized fork as a termination of replication intermediate in Bacillus subtilis, Journal of Molecular Biology, 188(2), 199-205.
| Crossref | Google Scholar | PubMed |
Wilce, J. A., Vivian, J. P., Hastings, A. F., Otting, G., Folmer, R. H., Duggin, I. G., Wake, R. G., and Wilce, M. C. (2001) Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest, Nature Structural Biology, 8(3), 206-210.
| Crossref | Google Scholar | PubMed |
Footnotes
12 Wake (1963).
59 Wake (1972)
25 Wake (1972).
26 Wake (1967).
28 Wake (1972). Wake (1973).
48 Wake (1996).
57 Duggin (2006).



