Dimethylsulfoniopropionate in corals and its interrelations with bacterial assemblages in coral surface mucus
P. R. Frade A B G , V. Schwaninger C , B. Glasl A , E. Sintes A , R. W. Hill D , R. Simó E and G. J. Herndl A FA University of Vienna, Department of Limnology and Bio-Oceanography, Althanstrasse 14, AT-1090 Vienna, Austria.
B Caribbean Research and Management of Biodiversity (CARMABI) Foundation, Piscaderabaai z/n, PO Box 2090, Willemstad, Curaçao.
C University of Innsbruck, Institute of Ecology, Technikerstrasse 25, AT-6020 Innsbruck, Austria.
D Michigan State University, Department of Zoology, 288 Farm Lane RM 203, East Lansing, MI 48824, USA.
E Institute of Marine Sciences (ICM-CSIC), Passeig Marítim de la Barceloneta 37-49, E-08003 Barcelona, Spain.
F Royal Netherlands Institute for Sea Research (NIOZ), Department of Biological Oceanography, 1790 AB Den Burg, Netherlands.
G Corresponding author. Email: pedro.rodrigues.frade@univie.ac.at
Environmental Chemistry 13(2) 252-265 https://doi.org/10.1071/EN15023
Submitted: 24 January 2015 Accepted: 3 April 2015 Published: 27 August 2015
Journal Compilation © CSIRO Publishing 2016 Open Access CC BY-NC-ND
Environmental context. Corals produce copious amounts of dimethylsulfoniopropionate (DMSP), a sulfur compound implicated in climate regulation. We studied DMSP concentrations inside corals and unveiled the linkage between DMSP availability and the abundance of DMSP-degrading bacterial groups inhabiting the corals’ surface. Our findings suggest that DMSP mediates the interplay between corals and microbes, highlighting the importance of sulfur compounds for microbial processes in corals and for the resilience of coral reef ecosystems.
Abstract. Corals produce copious amounts of dimethylsulfoniopropionate (DMSP), a sulfur compound thought to play a role in structuring coral-associated bacterial communities. We tested the hypothesis that a linkage exists between DMSP availability in coral tissues and the community dynamics of bacteria in coral surface mucus. We determined DMSP concentrations in three coral species (Meandrina meandrites, Porites astreoides and Siderastrea siderea) at two sampling depths (5 and 25 m) and times of day (dawn and noon) at Curaçao, Southern Caribbean. DMSP concentration (4–409 nmol cm–2 coral surface) varied with host species-specific traits such as Symbiodinium cell abundance, but not with depth or time of sampling. Exposure of corals to air caused a doubling of their DMSP concentration. The phylogenetic affiliation of mucus-associated bacteria was examined by clone libraries targeting three main subclades of the bacterial DMSP demethylase gene (dmdA). dmdA gene abundance was determined by quantitative Polymerase Chain Reaction (qPCR) against a reference housekeeping gene (recA). Overall, a higher availability of DMSP corresponded to a lower relative abundance of the dmdA gene, but this pattern was not uniform across all host species or bacterial dmdA subclades, suggesting the existence of distinct DMSP microbial niches or varying dmdA DMSP affinities. This is the first study quantifying dmdA gene abundance in corals and linking related changes in the community dynamics of DMSP-degrading bacteria to DMSP availability. Our study suggests that DMSP mediates the regulation of microbes by the coral host and highlights the significance of sulfur compounds for microbial processes in coral reefs.
Introduction
Dimethylsulfoniopropionate (DMSP), an organic sulfur molecule, plays a key role in marine biogeochemical and ecological processes.[1,2] DMSP is also produced in copious amounts by reef-building corals.[3–5] The long-standing paradigm of strict algal-based DMSP production[6] has recently been challenged by the corroboration of DMSP biosynthesis in coral animals lacking their phototrophic endosymbionts.[7] These symbionts, the so-called zooxanthellae (genus Symbiodinium), constitute a genetically diverse assemblage whose functional diversity confers their hosts an important path of adaptation to the surrounding environment, particularly concerning light harvesting and resilience to thermal stress.[8,9] The abundance of Symbiodinium within the tissue of coral hosts has been correlated with tissue DMSP concentrations among and within cnidarian species.[10] Irrespective of its source, DMSP produced by coral symbioses becomes available to coral-associated bacteria that may use this compound as a source of reduced sulfur and carbon, and metabolise it to dimethylsulfide (DMS),[11,12] a volatile gas often implicated in climate regulation.[13,14]
Degradation of DMSP by marine microorganisms takes place by at least two major pathways, mainly distinguished by the fate of sulfur and methyl groups.[11] The cleavage pathway involves degradation of DMSP to DMS by phytoplankton or bacteria using enzymes known as DMSP lyases, with the loss of sulfur and methyl groups by diffusion of DMS.[15,16] The alternative demethylation pathway, carried out by bacteria alone, involves an initial demethylation of DMSP to methylmercaptopropionate (MMPA), with some portion of the sulfur being assimilated into sulfur-containing amino acids such as methionine and cysteine.[1,17,18] The first step in the demethylation pathway involves a single gene, DMSP demethylase (dmdA), first discovered in the Roseobacter group,[19] and since then shown to be widespread in marine bacterial communities,[20] including coral-associated communities.[21] The relative incidence of these competing microbial pathways has important consequences for the marine sulfur cycle and for DMS release into the atmosphere.[2,22]
Besides its role as a carbon and sulfur source for bacteria, DMSP has been shown to be involved in osmoprotection in algae and bacteria,[6,23] to be a precursor of cues for chemosensory attraction for a variety of organisms from bacteria to vertebrates[24,25] and to be an antiviral defence mechanism.[26] Most importantly, DMSP and its enzymatic breakdown products, DMS, acrylate, dimethylsulfoxide (DMSO) and methane sulfinic acid (MSNA), are scavengers of hydroxyl radicals and other reactive oxygen species (ROS) in marine algae.[27] Hypothesised as an antioxidant also in coral symbioses,[28] DMSP is therefore ecologically relevant because it may help corals to survive conditions of thermal stress,[7] and because it likely plays a major role in structuring coral-associated bacterial communities.[21] Both phenomena might have significant consequences at the ecosystem level. Reef-building corals associate with dynamic and highly diverse consortia of bacteria,[29] particularly abundant at the interface between the coral host epithelium and the ambient seawater.[30] Here, surface mucus secreted by the host’s mucocytes offers a nutrient- and niche-rich microhabitat in which microbes thrive.[31] Functionally, mucus-associated microbes seem crucial to the physiology of their hosts by contributing to defence and resistance against pathogens and to biogeochemical cycling.[32] Microbial biofilms established on the surface mucus of corals are subjected to high concentrations of DMSP, which leaks from the coral tissue, and reaches concentrations within mucus 2–4 fold higher than in the surrounding reef water.[33] Thus, it is likely that there is substantial DMSP degradation taking place within the surface mucus layer of corals, mediated by members of the mucus microbiome. Thus far, dmdA gene abundance has not been reported for corals. As corals represent microhabitats rich in DMSP, it is crucial to study the coral-associated DMSP-demethylating microbial communities and to determine whether there are distinct DMSP microniches.
In the present study, we follow up on recent findings suggesting a structuring role for coral-produced DMSP on coral-associated bacterial communities.[21] We tested the hypothesis that there is a link between coral DMSP availability and the community dynamics of DMSP-demethylating bacteria inhabiting the surface mucus of such corals. We first determined in situ DMSP concentrations in the tissue of three common reef-building coral species in relation to the intensity of solar radiation and to desiccation stress caused by air exposure. We then used genetic analysis to investigate the taxonomic affiliation and relative abundance of mucus-associated bacterial assemblages harbouring the DMSP demethylase gene, aiming to elucidate the role of DMSP in shaping coral-associated microbial communities and, ultimately, its significance for microbial processes in coral reefs.
Experimental approach
Fieldwork and sample collection
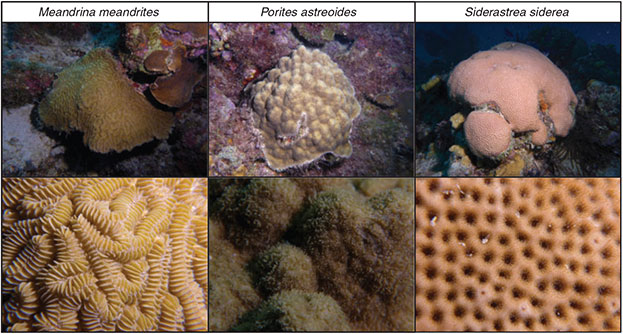
Fieldwork took place in March–April 2013 at Curaçao, former Netherlands Antilles. All samples were collected by scuba diving at the Buoy Zero reef, located ~500 m west of the Caribbean Research and Management of Biodiversity (CARMABI) Foundation’s field station (Fig. 1). The island of Curaçao is surrounded by a fringing reef, generally exhibiting a reef flat ~50–100 m wide, with a steep drop off starting at ~10–12-m depth, and another deep plateau at ~60-m depth.[34] Three common Caribbean reef-building coral species (see Fig. 2) were used in the current study: Meandrina meandrites (Linnaeus, 1758), Porites astreoides Lamarck, 1816 and Siderastrea siderea (Ellis and Solander, 1768). These species were chosen based on their high abundance at the study site in the form of fairly small colonies (~10–15 cm in diameter) easily detachable from the reef. Moreover, earlier research had demonstrated that DMSP occurs at a significantly high concentration in these species, possibly serving as an important osmolyte.[35] To investigate the role of light intensity on DMSP production, samples were taken at two different times of day (solar dawn and noon), and at two depths (5 and 25 m) to represent a gradient of solar radiation.[36] Sampling at noon was restricted to sunny days with minor cloud cover.
|
|
Samples for determining bacterial demethylase (dmdA) gene abundance within coral surface mucus were taken from undisturbed, healthy-looking coral colonies by gently rolling a sterile cotton swab over ~9–16 cm2 of their surface. Each swab was put inside a 2-mL microcentrifuge tube filled with air and held inverted (close to the sampled colony) to minimise contact of the swab with the surrounding water.
Coral tissue samples were taken by two approaches aimed at investigating the role of environmental stress on de novo DMSP production. A small skeleton chip covered by living tissue (~4 cm2 in area) was removed by hammer and chisel from approximately half of the abovementioned ‘swabbed’ colonies while they remained in situ on the reef. Chips of this type were considered ‘control’ samples. Each of them was placed immediately in an individual seawater-filled plastic bag protected from sunlight and quickly taken to the boat, where samples (including swab samples) were flash frozen in liquid nitrogen within 10–20 min of collection. The other half of the ‘swabbed’ colonies were collected as a whole, transported to the boat in individual plastic bags filled with seawater, and kept at constant temperature inside a cooling box. The colonies were then stressed by air exposure, providing our ‘stressed’ samples. Each colony was exposed to air for 3 min, immediately causing extensive (10–15 mL) mucus release from their surfaces. At that point, a small chip of each colony was removed and preserved as described for the control group. Air exposure of shallow-living corals, although not pervasive in the Caribbean, is a natural phenomenon occurring in other regions of the world, to which corals naturally respond by increasing mucus production. This is a basic adaptation to desiccation (among other environmental stresses) of all known corals. Upon return to the laboratory, all samples were kept at –80 °C until further processing, except while being transported to the University of Vienna, Austria, on dry ice.
Determination of DMSP and Symbiodinium abundances in coral tissue
In the laboratory of the Department of Limnology and Bio-Oceanography at the University of Vienna, coral chips were thawed and processed one at a time to reduce chances of sample degradation. Before thawing, samples were pre-washed with Milli-Q water (Merck Millipore, Darmstadt, Germany) to remove all traces of mucus and external seawater. The whole tissue associated with the coral chip was then blasted off using a jet of pre-chilled Milli-Q water and collected in a plastic bag. The blastate volume (100–300 mL) was determined, and subsamples for determining DMSP concentrations and zooxanthellae abundance were taken immediately after gentle homogenisation.
An 8-mL subsample was mixed 2 : 1 (v/v) with methanol and acidified to 3 % formic acid.[37] Acidification ensured that there was no chemical conversion of DMSP to DMS. Methanol was required for further analytical assays not reported here. All acidified subsamples were rapidly frozen in liquid nitrogen to minimise enzymatic cleavage of DMSP[38] and stored at –80 °C until they were shipped on dry ice to the Institute of Marine Sciences (CSIC) in Barcelona for DMSP determination. There, a total of 64 tissue blastate samples were analysed in duplicate by gas-phase chromatography using a flame photometric detector (GC-FPD, Shimadzu GC-14A, Duisburg, Germany). First, sodium hydroxide was added to the samples inside gas-tight vials without headspace. Aliquots (10–50 μL) were withdrawn with a gas-tight syringe through the vial septum and injected into the purge vial containing 2 mL of Milli-Q water. DMS evolved by alkaline hydrolysis was purged for 4–6 min with 40 mL min–1 of ultrahigh-purity helium (He), cryo-trapped in liquid nitrogen and subsequently snap volatilised at room temperature and injected into the GC-FPD. Calibration was performed by syringe injection into the purge vial of varying volumes of a gaseous mixture of He and DMS released by a weight-calibrated permeation tube (Dynacal, Valco Instruments Co. Inc.).[39] Plots of log(peak area) v. log(pmol DMS) yielded a straight line (R2 = 0.996) for < 35 pmols and a quadratic line (R2 = 0.998) for > 35 pmols, and were used for DMS quantification in the samples. Detection limit was 3 pmol and analytical precision was better than 10 %. Before running samples, tests with standard solutions (up to 26 μM) of DMS (Sigma–Aldrich, Steinheim, Germany) in methanol showed that there was no quenching effect of methanol on the sulfur signal. Because we did not measure residual DMS, we cannot exclude DMSP lyase activity during sample processing,[40] or incomplete oxidation of DMS to DMSO. Thus, residual DMS might have been quantified as DMSP by our method. However, throughout the text we will refer to DMSP only, because this is by far the more dominant compound in the coral system.[28]
Another subsample of the homogenised blastate was vortexed for 2 min to separate all zooxanthellae from host debris. Aliquots (1.5 mL) were then fixed in 0.5 % glutaraldehyde (final concentration, Sigma–Aldrich), stored in the dark for 10 min, flash frozen in liquid nitrogen and kept at –80 °C until further processing. Zooxanthellae abundances were estimated by flow cytometry (Accuri C6, BD Biosciences, Erembodegem, Belgium) on two aliquots. Counting events were kept within 200–1000 events per second by diluting the samples with TE buffer and all samples were run at a flow rate of 86 μL min–1. Zooxanthellae (Symbiodinium) were visible and gated as a distinct population of events with strong fluorescence signal and high forward scatter (proportional to cell diameter).[8] Symbiont abundances determined by flow cytometry (Accuri C6) were used to standardise DMSP concentrations per cell assuming the zooxanthellae are principally responsible for DMSP production within the coral symbiosis. For standardisation of DMSP relative to coral surface area, the bare skeleton chips resulting from the tissue blasting procedure were used. The surface area from which the homogenate originated was determined with the aluminium-foil method.[41] For M. meandrites, however, because of its meandroid morphology (see Fig. 2), this method was not considered adequate and an alternative approach using a calliper was applied. The surface area of skeletal septa was determined, their density over the skeleton estimated and a conversion between the aluminium foil and the calliper-geometry methods established, that was then applied to determine corrected surface areas of the skeletal chips. Areas of sampled fragments ranged between 2 and 45 cm2.
Nucleic acid extractions, dmdA gene amplifications and clone library generation
Mucus-saturated cotton tips of the swab samples were transferred to Lysing Matrix tubes of the Fast DNA Spin Kit for Soil (MP Biomedicals, Heidelberg, Germany) and disrupted in a FastPrep instrument (MP Biomedicals) for 40 s at a speed of 6.0. Further nucleic acid extraction followed the manufacturer’s protocol. Samples were eluted in 80 μL of desalinated (DES) water.
Clone libraries were generated for distinct dmdA gene assemblages[42] to determine the taxonomic affiliation and phylogenetic relations of mucus bacteria. Polymerase Chain Reaction (PCR) optimisation was initially conducted for several assemblages in dmdA clades A to E by testing a gradient of annealing temperatures on several samples. Successful PCR amplifications were obtained for four targeted dmdA subclades: A/2, C/2, D/1 and D/3, using specific quantitative PCR (qPCR) primers. Each 50-μL reaction consisted of 1× Taq buffer, 2 mM MgCl2, 0.2 mM of each deoxyribonucleotide (dNTP), 0.4 mg mL–1 BSA, 0.25 μM of each primer (see Table 1 for primer sequences), 0.02 U μL–1 of high fidelity Platinum taq polymerase (Invitrogen, Merelbeke, Belgium) and 4 μL of DNA extract. Thermo-cycling consisted of an initial denaturation step at 94 °C for 2 min; 42 cycles of amplification, with denaturing at 94 °C for 20 s; annealing at different temperatures (see Table 1 for details) for 30 s and extension at 72 °C for 30 s; and a final extension step at 72 °C for 15 min before holding reaction at 4 °C. All PCR products were checked on agarose gels after staining with SyBRGold (Molecular Probes, Invitrogen).

|
Clone libraries were generated for each of the four dmdA-gene subclades (see Table 1) for a single sample per species studied. The three samples included originated from the same depth (5 m) and time of the day (noon). For each sample and subclade, triplicate PCR reactions were pooled and purified using the PCR-Extract Mini Kit (5-Prime, Hilden, Germany). Purified PCR products were ligated into a TOPO-TA cloning vector (Invitrogen) and competent cells transformed following the manufacturer’s instructions. Transformants were selected on Luria Bertani agar plates (with 50 μg mL–1 amplicilin; Sigma–Aldrich) with X-gal (5-bromo-4-chloro-3-indolyl- β-D-galactopyranoside; Life Technologies, Darmstadt, Germany). White colonies were picked, re-inoculated on new agar plates, and immediately checked for inserts in a PCR with primers M13F (5′-GTA AAA CGA CGG CCA G-3′) and M13R (5′-CAG GAA ACA GCT ATG AC-3′). Clones that contained the correct dmdA gene fragment were then sent to GATC Biotech (Cologne, Germany) for sequencing (using the M13F primer), after being re-inoculated on agar plates. Obtained sequences were analysed with CodonCode Aligner (www.codoncode.com/aligner/, accessed 12 July 2013) and assembled sequences compared with BLASTX 2.2.30[43] to the refseq_protein database of the National Centre for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed 13 December 2014). Alignments of translated nucleotide sequences and Neighbour-Joining phylogenetic trees were generated in Geneious Pro 5.6 (www.geneious.com, accessed 14 December 2014)[44] using the Jukes-Cantor distance model. Sequence information has been deposited in GenBank under accession numbers KT069249-358 (for subclade A/2), KT069359–464 (for subclade C/2) and KT069465–549 (for subclade D/3)
Determination of relative abundance of bacterial dmdA gene assemblages by quantitative PCR
To determine the dmdA gene abundance of the dmdA-harbouring bacterial subclades (A/2, C/2, D/1 and D/3) qPCR was performed in polypropylene 96-well plates with optical sealing foils in a LightCycler 480 II (Roche, Rotkreuz, Switzerland). The recA gene, a house-keeping gene commonly used as the reference gene in qPCR studies of bacteria,[45] served to correct for possible variations in DNA extraction efficiencies between samples. Primers RECAF and RECAV (see Table 1) amplify a 212 bp region of the bacterial recA gene.[46] Each 10 μL reaction consisted of 1× LightCycler 480 SYBR Green I Mastermix (containing FastStart taq polymerase, reaction buffer, dNTP’s, SYBR Green I dye and MgCl2; Roche), 1 μM of each primer (see Table 1 for respective primers), and 3 μL of template DNA. For recA only 1 μL of template was used. Previous optimisation of the annealing temperatures for each specific primer set was conducted by running gradient PCRs in a standard thermocycler using the Roche Mastermix and the same cycling and temperature ramping conditions as in the qPCRs. Specificity of amplifications was at this point checked on 2 % agarose gels.
Standard curves were generated using the specific cloned dmdA subclades of known gene abundance. The DNA concentration was determined spectrophotometrically (Nanodrop, Thermo Fisher Scientific, Wilmington, DE, USA) after purification using the PCR-Extract Mini Kit (5-Prime) and the gene abundance calculated based on the fragment length and DNA concentration. Ten-fold dilutions of the standards in Tris buffer ranged between 101 and 107 targets μL–1. For recA, standard dilutions were prepared from the purified amplification product originating from a S. siderea sample. The slopes of the standard curves, correlation coefficients and amplification efficiencies of each primer pair were calculated using the analysis tools of the LC480 software release 1.5.0 (Roche).
The thermocycling protocol consisted of an initial denaturation step at 94 °C for 10 min, followed by 50 cycles of amplification, including denaturing at 94 °C for 30 s, annealing at the respective primer annealing temperature (see Table 1) for 40 s and extension at 72 °C for 30 s and 80 °C for 25 s (with readings taken at the end of each cycle). A melting curve analysis was performed to check for potential PCR artefacts by monitoring SYBR Green fluorescence in the temperature ramp of 60 to 94 °C with an increase of 0.5 °C and a hold for 1 s after each read. Products were finally stabilised at 4 °C for 15 min. All qPCR assays were conducted in triplicate for the standard, no-DNA controls and the environmental samples. The relative abundance of each subclade was calculated by dividing the number of the specific target gene by the recA gene abundance for each sample. This calculation assumes an equal number of dmdA gene copies for all subclades. Amplification efficiencies of qPCR are shown in Table 1.
Statistical analysis
All biological variables were analysed by Generalised Linear Modelling (GLM) to assess the significance of explanatory variables. Explanatory variables built into models were selected following biological reasoning. For instance, the variables ‘host species’, ‘depth’ and ‘time of the day’ were used as factors in models for the analysis of symbiont cell abundance. To build models for analysis of the variation in DMSP concentration, ‘symbiont cell abundance’ and ‘treatment’ (air exposure v. control) were introduced as factors, adding to the previous list of factors. Finally, to build models for analysis of bacterial dmdA gene abundance, the factors relating to ‘DMSP concentration’ were added on top of all the other factors mentioned above. Samples subjected to air exposure were excluded from these comparisons so that the putative effect of DMSP concentrations on dmdA gene abundance was not biased by the stress-related variation in DMSP. Thus, only the control group was included in statistical analysis of dmdA gene abundance. Pre-variable selection included a collinearity test based on the Variance Inflation Factor (VIF) statistic.[47] A VIF greater than 5 was considered an indicator of high colinearity and variables were excluded accordingly. Log-transformations were used to improve linearity and reduce outlier effect. Symbiont cell abundance and DMSP concentration were analysed by multiple linear regression (GLM using Gaussian distribution and identity function). Gene abundances generated by qPCR were analysed by assuming a Poisson distribution and using a log link function, an appropriate approach for counts originating from different units of collection. recA gene abundance was used as an offset variable in Poisson regression models to correct for the number of bacterial cells effectively extracted in each DNA sample. A backward and forward model selection tool was applied to identify the variables accounting for significant variation in each response variable. A model was considered adequate when no factor could be added or removed without causing a statistically significant change in the (residual) deviance (evaluated with the Akaike Information Criterion). The model was then validated by a nested model approach in which interaction effects were also tested. F statistics were used in linear regression and Chi-Square in log-linear models. In case of over-dispersion in Poisson models (deviance : d.f. residual > 150), the Quasi-poisson distribution was applied using the F-test for model selection and validation. All statistical tests were performed at a significance level of 0.01 and P-values adjusted according to Bonferroni correction. All statistical analyses were carried out using R statistics software (ver. 3.0.3, see www.r-project.org, accessed 14 December 2014). GLM analyses of variance tables are reported in the Supplementary material (Tables S1–S7). Throughout the manuscript, all reported values are mean ± s.d., unless mentioned otherwise.
Results
Sampling overview and Symbiodinium abundance in coral tissue
A total of 62 samples were collected, with a minimum of 6 colony replicates included in each sampling grouping (each species for each depth and time of the day). Owing to logistic limitations, no samples were collected from 25 m at dawn. Half of the total number of colonies were included in the air exposure experiment and constituted the ‘stressed group’ that comprised a total of 33 samples. The remaining 29 samples represented the ‘control group’, reflecting natural in situ conditions.
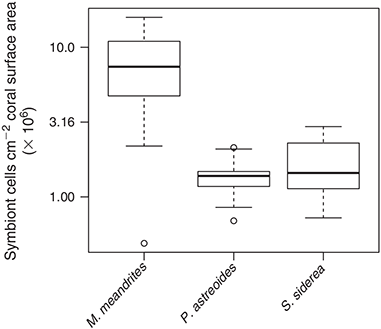
Symbiont cell abundance, determined by flow cytometry and normalised to coral surface area (n = 61), ranged between 0.5 and 15.9 × 106 cells cm–2 (Fig. 3), with a statistically significant effect of host species (F(2,58) = 51.136, P < 0.01). M. meandrites showed a higher symbiont abundance (7.9 ± 4.4 × 106 cells cm–2) than P. astreoides and S. siderea (1.4 ± 0.4 × 106 cells cm–2 and 1.7 ± 0.7 × 106 cells cm–2 respectively). No other explanatory variable was statistically related to symbiont abundance.
|
DMSP concentration in coral tissue
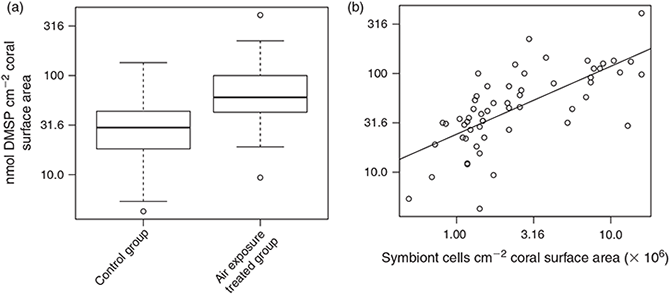
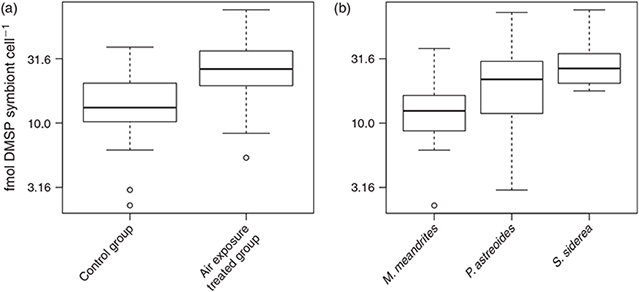
DMSP concentration (n = 57) ranged between 4 and 409 nmol cm–2 when normalised to coral surface area and between 2.3 and 76.2 fmol cell–1 when normalised to symbiont abundance. Tissue DMSP concentrations per unit area (Fig. 4) increased significantly under air exposure stress as compared with the control group (F(1,54) = 19.892, P < 0.01, Fig. 4a), and also increased significantly with increasing symbiont abundance in the tissue (F(1,54) = 62.641, P < 0.01, Fig. 4b). However, no statistical interaction effect was found between these two explanatory factors (F(1,53) = 0.129, P > 0.01). DMSP concentration per symbiont cell (Fig. 5) also increased significantly under air exposure stress as compared with control samples (F(1,53) = 13.541, P < 0.01, Fig. 5a), and varied significantly between host species (F(2,53) = 9.368, P < 0.01, Fig. 5b). No interaction effect between treatment and species was found (F(2,51) = 0.877, P > 0.01).
|
DMSP concentrations in natural in situ samples (control group) varied with the density of symbiont cells following the relationship:
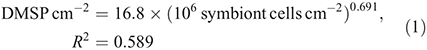
DMSP concentrations per symbiont cell (Fig. 5a) were higher in S. siderea (25.4 ± 8.2 fmol DMSP cell–1) than in M. meandrites (10.1 ± 4.8 fmol DMSP cell–1) and intermediate in P. astreoides (15.3 ± 8.5 fmol DMSP cell–1). The effect of air exposure stress led to a 2-fold increase of both DMSP per unit area (from 44.1 ± 42.4 in controls to 81.9 ± 74.5 nmol DMSP cm–2 in stressed colonies, Fig. 4a) and DMSP per symbiont cell (from 15.7 ± 9.1 in controls to 29.0 ± 16.7 fmol DMSP cell–1 in stressed colonies, Fig. 5a).
Coral mucus collected during the air exposure experiment exhibited DMSP concentrations ranging between 0.14 and 3.15 μM.
Phylogenetic affiliation of bacterial dmdA gene assemblages in coral mucus
A total of 389 clones were sequenced from the 12 dmdA gene clone libraries generated (for 3 coral species and 4 distinct bacterial dmdA subclades). The number of non-redundant sequences found was approximately one-tenth of all sequences. For clade D/1 no significant similarities were found in BLASTX searches because of the reduced length of the fragment. Figs S1, S2 and S3 depict phylogenetic reconstructions for dmdA subclades A/2, C/2 and D/3 respectively, including reference protein sequences with more than 80 % similarity to the translated sequences originated from the clone libraries. BLASTX searches for subclade A/2 yielded a large number of homologues, with the highest similarities found for Ruegeria sp. (97 %), Phaeobacter sp. (97 %), Roseobacter sp. (94 %), Silicibacter sp. (94 %) and Sulfitobacter sp. (94 %), all members of the class Alphaproteobacteria in the family Rhodobacteraceae. For dmdA subclade C/2 there was a low number of BLASTX hits. The only reasonably close hit to our translated sequences was Candidatus Pelagibacter ubique (87 % similarity), an abundant member of the SAR11 clade in the class Alphaproteobacteria. Finally, searches for subclade D/3 also yielded a reduced number of hits, of which the only one with a similarity above 80 % was Candidatus Pelagibacter ubique (with 88 %). The host species from which each sequence originated from did not resolve any particular topology within each of the dmdA gene trees (Figs S1–3). Overall, subclade A/2 appeared to contain substantially less sequence variability than subclades C/2 and D/3.
Relative abundance of bacterial dmdA gene assemblages in coral mucus
The dmdA gene abundance was determined by qPCR (see Table 1). All dmdA bacterial subclades showed qPCR specificity, with the exception of subclade D/1 that exhibited unspecific melting curves and very low fluorescence signal and was therefore excluded from further analysis. Several samples, however, were not quantifiable because the fluorescence signal was below the range covered by the respective standard serial dilutions.
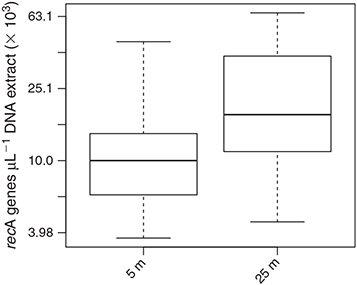
recA gene abundance ranged between 0.4 and 6.5 × 104 genes μL–1 of DNA extract (Fig. 6). recA gene abundance varied significantly between the two sampling depths (F(1,58) = 11.254, P < 0.01), with lower concentrations at 5 m (1.3 ± 1.0 × 104 recA genes μL–1) than at 25-m depth (2.5 ± 1.7 × 104 recA genes μL –1).
|
For subclade A/2, dmdA : recA ranged between 0.9 and 19.0 % (Fig. 7; n = 26) and varied significantly with DMSP concentration per unit area (χ2 = 559.0, d.f. = 1, P < 0.01), host species (χ2 = 3312.7, d.f. = 2, P < 0.01), time of collection (χ2 = 1221.9, d.f. = 1, P < 0.01) and sampling depth (χ2 = 34.0, d.f. = 1, P < 0.01). The effect of DMSP concentration was not constant across species (interaction: χ2 = 111.8, d.f. = 2, P < 0.01, Fig. 7a). The dmdA : recA ratio of the A/2 subclade (Fig. 7b, 7c) was higher in P. astreoides (6.0 ± 5.8 %) than in S. siderea (2.5 ± 1.4 %) and M. meandrites (1.8 ± 0.9 %); higher at noon (4.2 ± 4.6 %) than at dawn (1.9 ± 0.7 %); and higher at 25-m depth (3.9 ± 4.0 %) than at 5-m depth (3.4 ± 4.3 %). The dmdA : recA ratio of the A/2 subclade changed with DMSP concentration per unit area (Fig. 7a) according to the following equations:
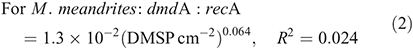
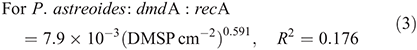
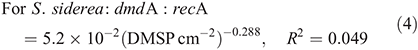
For subclade C/2, the dmdA : recA ratio ranged between 0.1 and 9.2 % (Fig. 8; n = 23) and varied significantly with DMSP concentration per unit area (χ2 = 108.4, d.f. = 1, P < 0.01), host species (χ2 = 2122.0, d.f. = 2, P < 0.01), time of collection (χ2 = 48.0, d.f. = 1, P < 0.01) and sampling depth (χ2 = 310.9, d.f. = 1, P < 0.01). Also for subclade C/2, the effect of DMSP concentration was not constant across species (interaction: χ2 = 46.5, d.f. = 2, P < 0.01, Fig. 8a). The dmdA : recA ratio in the C/2 subclade (Fig. 8b, c) was higher in P. astreoides (4.5 ± 2.8 %) than in M. meandrites (2.2 ± 3.3 %) and S. siderea (1.5 ± 1.3 %); higher at dawn (4.0 ± 3.0 %) than at noon (2.6 ± 2.8 %); and higher at 5 m (3.6 ± 3.0 %) than at 25 m (2.1 ± 2.7 %). The dmdA : recA ratio within the C/2 subclade changed with DMSP concentration per unit area (Fig. 8a) according to the following equations:
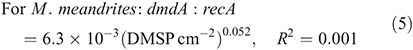
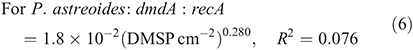
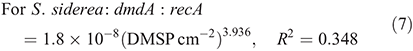
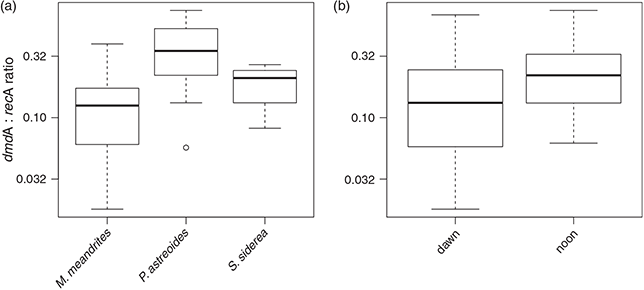
For subclade D/3, dmdA : recA ratios ranged between 1.8 and 74.5 % (Fig. 9; n = 22) and varied significantly with host species (F(2,18) = 23.883, P < 0.01) and time of collection (F(1,18) = 17.889, P < 0.01). The dmdA : recA ratio (Fig. 9a, b) in the D/3 subclade was higher in P. astreoides (37.6 ± 23.9 %) than in S. siderea (18.7 ± 7.8 %) and M. meandrites (14.2 ± 11.8 %); and higher at noon (26.6 ± 18.6 %) than at dawn (21.1 ± 23.7 %).
|
Discussion
DMSP concentration in coral tissue and de novo production
Since the first descriptions of DMSP in corals,[4,5] several studies have reported on the concentration of this metabolite in the tissue of different coral species. DMSP concentrations of 100–300 nmol DMSP cm–2 have been reported for several Hawaiian corals,[4] and concentrations ranging from nanomole to micromole per square centimetre were reported for the tissue of juvenile corals even in the absence of zooxanthellae.[7] DMSP concentrations per symbiont cell ranged between 20 and 4000 fmol DMSP cell–1 for corals of the Great Barrier Reef.[3] For the three Caribbean coral species analysed in the present study (see Fig. 2), preliminary data obtained by a different assay (liquid chromatography–mass spectrometry) and based on only a few samples reported averages of ~460 nmol cm–2, 113 nmol cm–2 and 40 nmol cm–2 for M. meandrites, P. astreoides and S. siderea respectively.[35] The DMSP concentrations we obtained ranging from 4–400 nmol DMSP cm–2 of coral surface and 2–80 fmol DMSP symbiont cell–1 are well within the range previously reported. Thus, we confirm the potential of corals as reservoirs for DMSP. Furthermore, our DMSP concentrations obtained for coral mucus (in the μM range) match those of Broadbent and Jones[33] for mucus of corals of the Great Barrier Reef (2–54 μM), that were 2–4 fold higher than those determined for the surrounding reef water.
A striking finding of our study was that stressed coral colonies (3 min of air exposure) showed a 2-fold increase in DMSP concentrations (Figs 4a, 5a). This rapid response suggests rapid de novo production of DMSP by the coral symbiosis, either by Symbiodinium or by the coral host itself,[7] probably triggered by the oxidative stress concomitant with irradiance or thermal stress.[48] This instant production of DMSP highlights the role of this molecule as a stress response osmolyte[6] and an antioxidant,[28] a conclusion supported by the great variation in DMSP concentrations when corals are recovering from stress events such as mass bleaching.[49] Recent studies have explicitly implicated DMSP and its enzymatic breakdown products in the stress response of corals, as scavengers of cellular reactive oxygen species (ROS) generated upon thermal stress[7,28] or upon direct addition of oxidative stressors such as copper.[50] These recent findings add to others postulating a protective role of DMSP and its degradation products dimethylsulfide (DMS), acrylate, dimethylsulfoxide (DMSO) and methane sulfinic acid (MSNA) as efficient scavengers of hydroxyl radicals and other ROS in free-living algae.[27,51]
The increase in DMSP upon air exposure was evident, independently of whether DMSP concentrations are normalised to coral surface or symbiont abundance. These two normalisation approaches, however, represent two different conceptual views, or at least, allow interpretation of different processes. The normalisation per unit area does not imply any assumption about the localisation of the DMSP within the tissue and rather stresses the link between the coral’s life history and metabolic processes mediated by the coral surface. The normalisation per symbiont cell, however, assumes that most of the DMSP is produced by Symbiodinium. The data obtained by normalisation of DMSP per unit area likely relate more closely to conditions in the surface mucus layer, because all DMSP released to the surrounding seawater has to diffuse through the mucus layer. In contrast, normalisation per symbiont cell would be a better approach to infer physiological processes, such as oxidative stress, taking place inside the tissue. Ultimately the two normalisations are related by a factor defined as the number of symbionts per unit coral surface area. This means their relatedness is disrupted when corals with distinct tissue thicknesses are compared. Although we do not have data on tissue thickness, our field observations suggest that M. meandrites has a thicker tissue than those of S. siderea and P. astreoides, therefore putatively explaining the specific differences observed in symbiont abundance (Fig. 3). Symbiont abundances (per coral surface area), higher in M. meandrites than in the other two species, are known to be a species-specific trait and probably related to three-dimensional space availability within the coral tissue[8] and to the genetically constrained cell size of particular symbiont lineages.[52] Although Symbiodinium numbers inside the coral tissue are known to vary over time and space for individual species,[36,53] neither time of the day nor depth explained their variation in the current study.
Approximately 60 % of the variation in DMSP concentration per unit area was, according to our multiple linear regression models, explained by symbiont abundance within the tissue (see Fig. 4b), a pattern also previously inferred by van Alstyne et al.[10] from comparisons among and within other four cnidarian species. Basically, a larger pool of DMSP-producing symbiont cells would mean a larger pool of DMSP. However, we cannot exclude that the host contributes, at least partly, to DMSP production.[7] Also, higher symbiont abundances correspond frequently to larger coral biomass (thicker tissue). Determination of DMSP concentration per symbiont cell, however, is not biased by tissue thickness or by surface area calculations, as both DMSP concentrations and cell abundance are measured from equivalent subsamples. Our analysis showed that DMSP concentrations per symbiont cell relate to the identity of the host (Fig. 5b), suggesting that there are species-specific differences in DMSP production or retention, that could either relate to the host itself or to genetic differences in the symbiont lineages harboured by these coral species. Actually, distinct Symbiodinium genetic lineages are known to exhibit different thermal tolerances.[9] Thermal stress has been shown to induce enhanced consumption of both DMS and DMSP in more thermo-sensitive Symbiodinium genotypes as compared with thermo-tolerant ones.[54] Therefore, the fact that M. meandrites hosts exclusively Symbiodinium type B1, P. astreoides hosts mixed Symbiodinium assemblages belonging to the A, B and C clades, and S. siderea hosts predominantly Symbiodinium type C3[55] could, per se, explain the observed patterns. A non-mutually exclusive alternative explanation is that the distinct symbiont lineages present in different hosts exhibit distinct DMSP lyase activities.[56]
Overall, our interpretation of the results implicates the occurrence of higher symbiont cell abundance as a cause for the higher amounts of DMSP in the tissue, and that the DMSP concentration per symbiont cell does not necessarily relate to DMSP release rates at the level of the coral’s surface. DMSP release rates are likely more dependent on tissue thickness and the abundance of symbionts per surface area. The absence of effect of time of the day or depth on DMSP concentration suggests that either DMSP production in coral symbioses is not directly related to light availability, that light-dependent processes of production and consumption of DMSP within corals balance each other, or that the coral host provides light modulation mechanisms capable of buffering the influence of external light availability on DMSP biosynthesis. An example of such a mechanism is the photoprotective regulation of the light environment within the coral tissue provided by tissue-associated fluorescent pigments.[57] Although we cannot exclude that the range of light irradiance in the current study was too narrow to cause significantly different levels of DMSP, colonies collected at 5 m experience far higher photosynthetic radiation (PAR) than at 25 m: ~1000 μmol photons m–2 s–1 of downwelling irradiance at 5 m v. 250 μmol photons m–2 s–1 at 25 m.[36]
In the study of Hill et al.,[35] focussing on the photoprotective role of betaines (quaternary ammonium metabolites) in corals, DMSP was also included because of its related chemical structure (a tertiary sulfonium compound). Consistent with our results, Hill et al.[35] found that DMSP concentrations of members of the genus Madracis did not vary significantly with irradiance exposure (after comparisons between different depths, between shaded and exposed colonies and between different times of the day). Together with the rapid response to air exposure we reported, these findings suggest that the mechanisms of DMSP up-regulation are easily masked by DMSP down-regulation (lyase activity, for instance) for much of the time, even for colonies living under very different light regimes, and that only upon extreme stress does DMSP production become much higher than its consumption. Air exposure and thermal stress[7] are examples of such extreme stresses.
Phylogenetic affiliation of bacterial dmdA gene assemblages in coral mucus
Demethylase gene sequences generated in this study all closely affiliate to Alphaproteobacteria, an expected finding considering that most bacterial taxa known to be involved in DMSP demethylation belong to this class.[20] The only dmdA gene clade so far known to relate to other bacterial groups, clade E (Gammaproteobacteria), was not detected in preliminary amplifications and was therefore not further investigated in qPCR assays. Although initial PCR amplifications targeting other dmdA clades and subclades described by Varaljay et al.[42] were not successful, this does not necessarily mean that they are absent from the mucus microbiome. Clade A is known to represent marine Roseobacter and Rhodospirillales species, whereas clade B includes the only sequenced marine SAR116 group member. Actually, to date, nearly all known DMSP-catabolising bacteria belong to the phylum Proteobacteria.[58] Within the Alphaproteobacteria there were, however, striking differences detectable, with dmdA subclades C/2 and D/3 affiliating closely with members of the family Pelagibacteraceae, whereas subclade A/2 was closely related to a diverse assemblage of taxa belonging to the family Rhodobacteraceae. Roseobacters are a group of abundant marine bacteria known to demethylate DMSP, and Pelagibacter belongs to the SAR11 clade, whose members are highly dominant organisms in marine ecosystems, that are also well known to demethylate DMSP[11] and for which the dmdA protein structure is described.[59]
The low sequence variation detected in our clone libraries, and the fact that a large fraction of bacterial dmdA taxonomic diversity was shared between all three coral host species sampled, suggest the absence of endemic dmdA assemblages or strict species-specific coral niches for DMSP demethylating bacteria within the surface mucus layer of corals. A recent study comparing free-living v. particle-associated bacterial dmdA assemblages found that most dmdA genes were shared between these two habitats,[42] again suggesting no niche diversification of dmdA gene assemblages towards distinct habitats. The rather uniform G+C content of dmdA genes led Howard et al.[20] to propose that lateral gene transfer may have contributed to the wide distribution of dmdA and its uniformity among diverse taxa. However, these conclusions do not preclude the existence of distinct bacterial DMSP niches. The differences we found by quantifying distinct dmdA gene assemblages (Figs 7–9) indicate that some niche differentiation does occur for bacterial DMSP demethylation.
Relative abundance of bacterial dmdA gene assemblages in coral mucus
We hypothesised that there is a link between DMSP availability and the relative abundance of DMSP-demethylating bacteria. The dmdA : recA gene abundance ratios we obtained suggest that a large proportion of the mucus-associated bacterial community actually harbours demethylase genes (Figs 7–9). Although we did not quantify all dmdA gene subclades described by Varaljay et al.,[42] the sum of the three dmdA clades adds up to almost 100 % of the recA gene abundance, suggesting that the majority of the bacterial species associated with corals are involved in DMSP degradation as also indicated by Raina et al.[21]
The presence of DMSP demethylase genes in coral mucus is an indicator of the potential for the DMSP demethylation pathway that, if active, ultimately leads to the production of the highly reactive sulfur species methanethiol (MeSH) constituting an alternative pathway to the DMS-producing cleavage pathway.[11] Thus, DMSP demethylation routes the sulfur moiety away from the climatically active dimethylsulfide, ultimately shunting part of the sulfur to sulfur-containing amino acids such as methionine.[1,17,18] This bacterial switch between producing DMS or MeSH[1] has consequences for bacterial growth and has been hypothesised to be controlled by DMSP availability.[2] This hypothesis suggests that when DMSP availability is low (or there is high bacterial sulfur demand), a higher fraction of DMSP is channelled into demethylation and assimilation. This poses an interesting theoretical background to interpret shifts in bacterial dynamics linked to DMSP availability. So far, however, only inconclusive evidence exists for the connection between DMSP dynamics and changes in DMSP assimilating bacterial community composition. Howard et al.,[60] for instance, conjectured that differential growth rates among bacterioplankton taxa and shifts in their taxonomic composition after a phytoplankton bloom might be related to DMSP dynamics. More direct links have been established at the behavioural level, with DMSP as the main agent promoting chemotactic behaviour of a known coral pathogen towards the mucus of its host.[25]
Community level changes in coral-associated bacteria in response to DMSP have not been shown thus far. From the three dmdA subclades examined, only D/3 did not appear to respond to DMSP concentrations. In contrast, dmdA subclades A/2 and C/2 suggest that DMSP might play a role in structuring mucus-associated bacterial assemblages. Nevertheless, the bacterial community response to DMSP, as estimated by changes in the relative abundance of demethylase gene assemblages, is not uniform among host species or bacterial subclades (see Figs 7a, 8a). Overall, in P. astreoides, the species with the lowest concentration in DMSP, dmdA gene abundance is positively related to DMSP concentration for both subclades A/2 and C/2. For M. meandrites, the species with the highest concentration in DMSP, there was no relation of dmdA gene abundance with DMSP concentration for any of the three clades. In S. siderea, the coral species with low to intermediate levels of DMSP, contrasting results were obtained between dmdA subclades A/2 and C/2, with the former decreasing in abundance and the latter increasing in abundance with increasing DMSP concentration. A closer analysis of the data patterns for subclade D/3 suggests that if the detected overdispersion (leading to a Quasipoisson distribution) had been ignored, the response of dmdA gene abundance to DMSP concentration would mimic that of subclade C/2. This is an interesting note because both subclades C/2 and D/3 are phylogenetically associated with the family Pelagibacteraceae, whereas subclade A represents Rhodobacteraceae.
It is difficult to determine whether the reported differences in the relative abundance of the distinct dmdA subclades are related to changes in overall bacterial abundance. Although recA abundance was used as a proxy for bacterial abundance in the different samples (because there is a single copy of the recA gene per bacterial genome and because the same DNA extraction procedure was applied to all samples), the only factor influencing the variation of recA gene abundance was depth (Fig. 6). One could therefore assume that there is a higher bacterial abundance in the surface mucus of corals living at 25-m depth than at 5-m depth, and that the higher dmdA : recA ratio measured for subclade A/2 at 25 m relates to favourable bacterial growth of this subclade relative to that of subclade C/2. Nevertheless, because there is no hard evidence for this inference, our interpretation focuses only on differences of relative abundance between dmdA subclades. Interestingly, samples with dmdA gene abundance below qPCR detection limits always had a low recA abundance (<5000 recA genes μL–1 of DNA extract), suggesting that these low signals are not necessarily relating to low relative abundance of dmdA harbouring bacteria but rather to an overall low absolute bacterial abundance.
Assuming that changes in the relative abundance of dmdA genes relate to the degree of adaptation of the respective subclades to their environment, and assuming that the putative metabolic pathways of these subclades would be constrained or determined by changes in the carbon and sulfur mass balance mediated by demethylation of DMSP, one can speculate on a scenario in which DMSP availability[2] and DMSP affinity may explain these general trends. Fig. 10 depicts this explanation. With increasing DMSP concentrations at low initial concentrations the capacity to utilise DMSP in demethylation increases until a saturation level. At DMSP concentrations above this saturation level, demethylation rates are not further increasing independently of the DMSP concentration. At high DMSP concentrations the less costly pathway of DMSP cleavage is likely used,[11] contributing to the success of other groups of bacteria, that would then become more abundant. Demethylation likely quickly gives way to DMSP cleavage as the substrate becomes more abundant[2] until a point is reached where rates of DMSP lyase activity are not increasing anymore. In this conceptual model, dmdA subclade A/2 is hypothesised to have a higher affinity for DMSP than C/2, therefore explaining the difference between the positive and negative dmdA-DMSP relationships observed in S. siderea.

|
Although little is known about the kinetic properties of dmdA enzymes, Candidatus Pelagibacter ubique and Ruegeria pomeroyi have similarly low affinities for DMSP with a Km of 13.2 and 5.4 mM respectively.[61] Curiously, Ruegeria pomeroyi, with a lower Km (higher substrate affinity) is a close relative to the sequences we retrieved from subclade A/2, the dmdA assemblage we speculate has the highest affinity to low concentrations of DMSP. Candidatus Pelagibacter ubique is, in contrast, a close relative to subclade C/2.
Because our analysis only encompassed a limited fraction of the mucus-associated bacterial community, it is possible that different trends would emerge when including other dmdA gene clusters. Comparing the taxonomic affiliations of the dmdA gene assemblages described in the present study with those obtained from a 16S rRNA gene amplicon high-throughput sequencing analysis currently being carried out for the mucus microbiomes of the same coral species (P. R. Frade, unpubl. data), reveals that our qPCR approach has targeted a maximum of ~10 % of the total number of microbes living in the coral mucus layer (Rhodobacteraceae represents ~1–4 % and Pelagibacteraceae ~3–6 % of all sequences generated). We found patterns linking DMSP concentration to abundance of dmdA gene assemblages suggesting that a range of DMSP microniches are present in corals. It is possible, however, that such patterns are the result of taxonomic shifts dependent on other factors such as specificity of bacterial communities towards the host[62] or the symbiont populations harboured by the host,[63] or even a response to other molecules varying concomitantly with DMSP. The fact that we found DMSP-dmdA linkages both within and among coral species supports our ecological interpretation of the results. Further studies on coral-associated communities capable of demethylating DMSP are required to test the validity of the conceptual model illustrated in Fig. 10.
Concluding remarks
This is the first study to provide quantification of dmdA gene assemblages in corals and to link related changes in the community dynamics of DMSP-degrading bacteria to the availability of DMSP. This linkage highlights the important metabolic bridge established by DMSP between the coral host (with its phototrophic endosymbionts) and its associated microbial communities, and therefore confirms DMSP as an important molecule mediating the interplay between eukaryotes and prokaryotes. Our findings contribute to the established research field on the ecological roles of DMSP, to the emergent study of microbial mediated DMSP/DMS dynamics[64] and, in the larger context, to the understanding of multi-trophic level mutualisms in marine ecosystems.[65]
Acknowledgements
Thanks to Catarina Silva for logistical assistance provided during fieldwork and Christian Baranyi for laboratory assistance at the University of Vienna. Financial support to the first author was provided by a Marie Curie fellowship from the European Commission (project FP7-299320) and a fellowship from the Lise Meitner Program of the Austrian Science Fund (FWF, project M1363-B20).
References
[1] R. Simó, Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 2001, 16, 287.| Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links.Crossref | GoogleScholarGoogle Scholar | 11369106PubMed |
[2] R. P. Kiene, L. J. Linn, J. A. Bruton, New and important roles for DMSP in marine microbial communities. J. Sea Res. 2000, 43, 209.
| New and important roles for DMSP in marine microbial communities.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXms1Wrtbw%3D&md5=789cb96d8406d5bbead908e442a540f1CAS |
[3] A. D. Broadbent, G. B. Jones, R. J. Jones, DMSP in corals and benthic algae from the Great Barrier Reef. Estuar. Coast. Shelf Sci. 2002, 55, 547.
| DMSP in corals and benthic algae from the Great Barrier Reef.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xot1Kmsbs%3D&md5=070e582236c54ed605e704f487a7d2d9CAS |
[4] R. W. Hill, J. W. H. Dacey, D. A. Krupp, Dimethylsulfoniopropionate in reef corals. Bull. Mar. Sci. 1995, 57, 489.
[5] G. B. Jones, M. A. J. Curran, A. D. Broadbent, Dimethylsulphide in the South Pacific, in Recent Advances in Marine Science and Technology ‘94: Sixth Pacific Congress on Marine Science and Technology (Eds O. Bellwood, H. Choat, N. Saxena) 1994. p. 183–190 (PACON International and James Cook University of North Queensland: Townsville, Qld).
[6] J. Stefels, Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 2000, 43, 183.
| Physiological aspects of the production and conversion of DMSP in marine algae and higher plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXms1Wrtb4%3D&md5=23897818cf9950bd364d15554611ea98CAS |
[7] J. B. Raina, D. M. Tapiolas, S. Foret, A. Lutz, D. Abrego, J. Ceh, F. O. Seneca, P. L. Clode, D. G. Bourne, B. L. Willis, C. A. Motti, DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 2013, 502, 677.
| DMSP biosynthesis by an animal and its role in coral thermal stress response.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs12ksL%2FN&md5=54e55bace7bdd8180dd2edcf2cee3f33CAS | 24153189PubMed |
[8] P. R. Frade, P. Bongaerts, A. J. S. Winkelhagen, L. Tonk, R. P. M. Bak, In situ photobiology of corals over large depth ranges: a multivariate analysis on the roles of environment, host, and algal symbiont. Limnol. Oceanogr. 2008, 53, 2711.
| In situ photobiology of corals over large depth ranges: a multivariate analysis on the roles of environment, host, and algal symbiont.Crossref | GoogleScholarGoogle Scholar |
[9] D. Tchernov, M. Y. Gorbunov, C. de Vargas, S. N. Yadav, A. J. Milligan, M. Haggblom, P. G. Falkowski, Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc. Natl. Acad. Sci. USA 2004, 101, 13531.
| Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXnvFeiu7k%3D&md5=e2d4f7f38233280a246d2834b5f8df38CAS | 15340154PubMed |
[10] K. L. Van Alstyne, P. Schupp, M. Slattery, The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates. Coral Reefs 2006, 25, 321.
| The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates.Crossref | GoogleScholarGoogle Scholar |
[11] C. R. Reisch, M. A. Moran, W. B. Whitman, Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2011, 2, 172.
| Bacterial catabolism of dimethylsulfoniopropionate (DMSP).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjvFWmtQ%3D%3D&md5=2a7361dbb94fc7c80ba7b2886bfdbf19CAS | 21886640PubMed |
[12] J. B. Raina, D. Tapiolas, B. L. Willis, D. G. Bourne, Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 2009, 75, 3492.
| Coral-associated bacteria and their role in the biogeochemical cycling of sulfur.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXntlShu74%3D&md5=37f0d05badc8e1574e35f123606e5c60CAS | 19346350PubMed |
[13] S. M. Vallina, R. Simó, Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 2007, 315, 506.
| Strong relationship between DMS and the solar radiation dose over the global surface ocean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXotFCisg%3D%3D&md5=89cd9bc46fb8728247c131f4a427b8cdCAS | 17255509PubMed |
[14] G. P. Ayers, J. L. Gras, Seasonal relationship between cloud condensation nuclei and aerosol methanesulfonate in marine air. Nature 1991, 353, 834.
| Seasonal relationship between cloud condensation nuclei and aerosol methanesulfonate in marine air.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXmvVCjs7g%3D&md5=5bafdf56ff9a700e243d744f6d995e12CAS |
[15] A. R. J. Curson, M. J. Sullivan, J. D. Todd, A. W. B. Johnston, DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 2011, 5, 1191.
| DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnvVaqsb0%3D&md5=97d019804bcf8b2321fb9a4d7eb5f217CAS |
[16] M. P. de Souza, D. C. Yoch, Purification and characterization of dimethylsulfoniopropionate lyase from an alcaligenes-like dimethyl sulfide-producing marine isolate. Appl. Environ. Microbiol. 1995, 61, 21.
| 1:CAS:528:DyaK2MXivValtb0%3D&md5=5b66324a297ac81e57c3b01bd9b4805dCAS | 16534905PubMed |
[17] R. P. Kiene, L. J. Linn, J. Gonzalez, M. A. Moran, J. A. Bruton, Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 1999, 65, 4549.
| 1:CAS:528:DyaK1MXms1Oitro%3D&md5=c0f9dc9c3f8951b78d1d03b07617b57dCAS | 10508088PubMed |
[18] R. P. Kiene, Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 1996, 54, 69.
| Production of methanethiol from dimethylsulfoniopropionate in marine surface waters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XlsFags78%3D&md5=b888cca6a19388bf5c6208323717afcaCAS |
[19] E. C. Howard, J. R. Henriksen, A. Buchan, C. R. Reisch, H. Burgmann, R. Welsh, W. Ye, J. M. González, K. Mace, S. B. Joye, R. P. Kiene, W. B. Whitman, M. A. Moran, Bacterial taxa that limit sulfur flux from the ocean. Science 2006, 314, 649.
| Bacterial taxa that limit sulfur flux from the ocean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFeitr7M&md5=d02b3f54b8d92d215347f6b84a76dc59CAS | 17068264PubMed |
[20] E. C. Howard, S. Sun, E. J. Biers, M. A. Moran, Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 2008, 10, 2397.
| Abundant and diverse bacteria involved in DMSP degradation in marine surface waters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFSksLvF&md5=b75790dff33d6b97cfc65d9c4ae98f90CAS | 18510552PubMed |
[21] J. B. Raina, E. A. Dinsdale, B. L. Willis, D. G. Bourne, Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 2010, 18, 101.
| Do the organic sulfur compounds DMSP and DMS drive coral microbial associations?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXjtVCksbs%3D&md5=bd9e73029ee5e8ff3fcd35a395a9f7feCAS | 20045332PubMed |
[22] M. Vila-Costa, R. Simó, H. Harada, J. M. Gasol, D. Slezak, R. P. Kiene, Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 2006, 314, 652.
| Dimethylsulfoniopropionate uptake by marine phytoplankton.Crossref | GoogleScholarGoogle Scholar | 17068265PubMed |
[23] D. T. Welsh, Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 2000, 24, 263.
| Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXjslOru7Y%3D&md5=358a169cf656df1eccfdd8fb755def5fCAS | 10841973PubMed |
[24] J. R. Seymour, R. Simó, T. Ahmed, R. Stocker, Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 2010, 329, 342.
| Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXosl2iu70%3D&md5=9160e8f0cec3e1f870bdb4bc1f5ff2baCAS | 20647471PubMed |
[25] M. Garren, K. Son, J. B. Raina, R. Rusconi, F. Menolascina, O. H. Shapiro, J. Tout, D. G. Bourne, J. R. Seymour, R. Stocker, A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 2014, 8, 999.
| A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXmvV2htbg%3D&md5=5b7327783a485908fcfa92b4b150727fCAS | 24335830PubMed |
[26] C. Evans, G. Malin, W. H. Wilson, P. S. Liss, Infectious titers of Emiliania huxleyi virus 86 are reduced by exposure to millimolar dimethyl sulfide and acrylic acid. Limnol. Oceanogr. 2006, 51, 2468.
| Infectious titers of Emiliania huxleyi virus 86 are reduced by exposure to millimolar dimethyl sulfide and acrylic acid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtVOisr7I&md5=e9fa0d1d17c5f4e80e3ac2c27346784dCAS |
[27] W. Sunda, D. J. Kieber, R. P. Kiene, S. Huntsman, An antioxidant function for DMSP and DMS in marine algae. Nature 2002, 418, 317.
| An antioxidant function for DMSP and DMS in marine algae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XltlGms7k%3D&md5=1bcaa8d9ca44d13d4f343901c31cbed2CAS | 12124622PubMed |
[28] E. S. M. Deschaseaux, G. B. Jones, M. A. Deseo, K. M. Shepherd, R. P. Kiene, H. B. Swan, P. L. Harrison, B. D. Eyre, Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnol. Oceanogr. 2014, 59, 758.
| Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtVKjt7zE&md5=b1629dff25674b8132acc86857455f2aCAS |
[29] T. D. Ainsworth, R. V. Thurber, R. D. Gates, The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 2010, 25, 233.
| The future of coral reefs: a microbial perspective.Crossref | GoogleScholarGoogle Scholar | 20006405PubMed |
[30] M. Garren, F. Azam, New method for counting bacteria associated with coral mucus. Appl. Environ. Microbiol. 2010, 76, 6128.
| New method for counting bacteria associated with coral mucus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtlartr3L&md5=5d3ef122035d6ceee1b24291a6eff453CAS | 20656857PubMed |
[31] J. C. Bythell, C. Wild, Biology and ecology of coral mucus release. J. Exp. Mar. Biol. Ecol. 2011, 408, 88.
| Biology and ecology of coral mucus release.Crossref | GoogleScholarGoogle Scholar |
[32] M. E. Mouchka, I. Hewson, C. D. Harvell, Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 2010, 50, 662.
| Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts.Crossref | GoogleScholarGoogle Scholar | 21558231PubMed |
[33] A. D. Broadbent, G. B. Jones, DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Mar. Freshwater Res. 2004, 55, 849.
| DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVens7jE&md5=74f3b38039bb5ef609d9ba545fa827e1CAS |
[34] R. P. M. Bak, Coral reefs and their zonation in the Netherlands Antilles. Stud. Geol. 1977, 4, 3.
[35] R. W. Hill, C. Li, A. D. Jones, J. P. Gunn, P. R. Frade, Abundant betaines in reef-building corals and ecological indicators of a photoprotective role. Coral Reefs 2010, 29, 869.
| Abundant betaines in reef-building corals and ecological indicators of a photoprotective role.Crossref | GoogleScholarGoogle Scholar |
[36] P. R. Frade, F. De Jongh, F. Vermeulen, J. Van Bleijswijk, R. P. M. Bak, Variation in symbiont distribution between closely related coral species over large depth ranges. Mol. Ecol. 2008, 17, 691.
| Variation in symbiont distribution between closely related coral species over large depth ranges.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXis1amtr0%3D&md5=7c45f6f75495e02cb597127ff04d8de0CAS | 18179427PubMed |
[37] C. Li, R. W. Hill, A. D. Jones, Determination of betaine metabolites and dimethylsulfoniopropionate in coral tissues using liquid chromatography-time-of-flight mass spectrometry and stable isotope-labeled internal standards. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010, 878, 1809.
| Determination of betaine metabolites and dimethylsulfoniopropionate in coral tissues using liquid chromatography-time-of-flight mass spectrometry and stable isotope-labeled internal standards.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnvV2qtLo%3D&md5=4956053263e5bfcdcd92be4dd66ed5d7CAS | 20627732PubMed |
[38] R. P. Kiene, D. Slezak, Low dissolved DMSP concentrations in seawater revealed by small-volume gravity filtration and dialysis sampling. Limnol. Oceanogr. Methods 2006, 4, 80.
| Low dissolved DMSP concentrations in seawater revealed by small-volume gravity filtration and dialysis sampling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XmtlWnt70%3D&md5=d76bee9995b6021df822bbc9b36611e1CAS |
[39] R. Simó, Trace chromatographic analysis of dimethyl sulfoxide and related methylated sulfur compounds in natural waters. J. Chromatogr. A 1998, 807, 151.
| Trace chromatographic analysis of dimethyl sulfoxide and related methylated sulfur compounds in natural waters.Crossref | GoogleScholarGoogle Scholar | 9646493PubMed |
[40] R. P. Kiene, D. J. Kieber, D. Slezak, D. A. Toole, D. A. del Valle, J. Bisgrove, J. Brinkley, A. Rellinger, Distribution and cycling of dimethylsulfide, dimethylsulfoniopropionate, and dimethylsulfoxide during spring and early summer in the Southern Ocean south of New Zealand. Aquat. Sci. 2007, 69, 305.
| Distribution and cycling of dimethylsulfide, dimethylsulfoniopropionate, and dimethylsulfoxide during spring and early summer in the Southern Ocean south of New Zealand.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1ajtrvK&md5=eaf7594ad6cad129fb5e6c65bc81eab1CAS |
[41] J. A. Marsh, Primary productivity of reef-building calcareous red algae. Ecology 1970, 51, 255.
| Primary productivity of reef-building calcareous red algae.Crossref | GoogleScholarGoogle Scholar |
[42] V. A. Varaljay, E. C. Howard, S. Sun, M. A. Moran, Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl. Environ. Microbiol. 2010, 76, 609.
| Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXksVCitLc%3D&md5=016ce7a8dcf10fe9f58c6828e0e3d9f2CAS | 19948858PubMed |
[43] S. F. Altschul, T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, D. J. Lipman, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389.
| Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXlvFyhu7w%3D&md5=affae6d90453d87dde7d1ceee8ad38e5CAS | 9254694PubMed |
[44] M. Kearse, R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, A. Cooper, S. Markowitz, C. Duran, T. Thierer, B. Ashton, P. Meintjes, A. Drummond, Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar | 22543367PubMed |
[45] G. W. Takle, I. K. Toth, M. B. Brurberg, Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 2007, 7, 50.
| Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum.Crossref | GoogleScholarGoogle Scholar | 17888160PubMed |
[46] D. E. Holmes, K. P. Nevin, D. R. Lovley, Comparison of 16S rRNA, nifD, recA, gyrB, rpoB and fusA genes within the family Geobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1591.
| Comparison of 16S rRNA, nifD, recA, gyrB, rpoB and fusA genes within the family Geobacteraceae fam. nov.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXptVOlsb0%3D&md5=96d9c481fb271b620fd8ff030102ee4bCAS | 15388715PubMed |
[47] D. C. Montgomery, E. A. Peck, Introduction to Linear Regression Analysis, 2nd edn 1992 (Wiley: New York).
[48] M. P. Lesser, Exposure of symbiotic dinoflagellates to elevated temperatures and ultraviolet radiation causes oxidative stress and inhibits photosynthesis. Limnol. Oceanogr. 1996, 41, 271.
| Exposure of symbiotic dinoflagellates to elevated temperatures and ultraviolet radiation causes oxidative stress and inhibits photosynthesis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xjs1eru7k%3D&md5=abc6b0d26cb7ea3240e3926b6c3b3741CAS |
[49] G. B. Jones, E. Fischer, E. S. M. Deschaseaux, P. L. Harrison, The effect of coral bleaching on the cellular concentration of dimethylsulphoniopropionate in reef corals. J. Exp. Mar. Biol. Ecol. 2014, 460, 19.
| The effect of coral bleaching on the cellular concentration of dimethylsulphoniopropionate in reef corals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXht1ensbzF&md5=d8a4bf603e1454ae99c8d59718611fcfCAS |
[50] D. M. Yost, R. J. Jones, C. L. Mitchelmore, Alterations in dimethylsulfoniopropionate (DMSP) levels in the coral Montastraea franksi in response to copper exposure. Aquat. Toxicol. 2010, 98, 367.
| Alterations in dimethylsulfoniopropionate (DMSP) levels in the coral Montastraea franksi in response to copper exposure.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmsVegtrw%3D&md5=502040f60fd24146627d6c679dc01876CAS | 20378188PubMed |
[51] D. Slezak, G. J. Herndl, Effects of ultraviolet and visible radiation on the cellular concentrations of dimethylsulfoniopropionate (DMSP) in Emiliania huxleyi (strain L). Mar. Ecol. Prog. Ser. 2003, 246, 61.
| Effects of ultraviolet and visible radiation on the cellular concentrations of dimethylsulfoniopropionate (DMSP) in Emiliania huxleyi (strain L).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXivVCjsLs%3D&md5=722f3b42da5bb8c59a44133ebd383b83CAS |
[52] T. C. LaJeunesse, G. Lambert, R. A. Andersen, M. A. Coffroth, D. W. Galbraith, Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 2005, 41, 880.
| Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVCksLrI&md5=60fa7d03b23d9ad02be383c33aee4e5dCAS |
[53] D. W. Kemp, X. Hernandez-Pech, R. Iglesias-Prieto, W. K. Fitt, G. W. Schmidt, Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 2014, 59, 788.
| Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtVKjt73O&md5=f1a9c5c9cfc0a38006a2d2d8288c534eCAS |
[54] E. S. M. Deschaseaux, V. H. Beltran, G. B. Jones, M. A. Deseo, H. B. Swan, P. L. Harrison, B. D. Eyre, Comparative response of DMS and DMSP concentrations in Symbiodinium clades C1 and D1 under thermal stress. J. Exp. Mar. Biol. Ecol. 2014, 459, 181.
| Comparative response of DMS and DMSP concentrations in Symbiodinium clades C1 and D1 under thermal stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtFWgtrnN&md5=bcbd28f2f69e9c1dc342c5f29eee514dCAS |
[55] P. Bongaerts, M. Carmichael, K. B. Hay, L. Tonk, P. R. Frade, O. Hoegh-Guldberg, Prevalent endosymbiont zonation shapes the depth distributions of scleractinian coral species. R. Soc. Open Sci. 2015, 2, 140297.
| Prevalent endosymbiont zonation shapes the depth distributions of scleractinian coral species.Crossref | GoogleScholarGoogle Scholar | 26064597PubMed |
[56] D. M. Yost, C. L. Mitchelmore, Dimethylsulfoniopropionate (DMSP) lyase activity in different strains of the symbiotic alga Symbiodinium microadriaticum. Mar. Ecol. Prog. Ser. 2009, 386, 61.
| Dimethylsulfoniopropionate (DMSP) lyase activity in different strains of the symbiotic alga Symbiodinium microadriaticum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVClu7bO&md5=c662707482021a361c975f01e0c51842CAS |
[57] A. Salih, A. Larkum, G. Cox, M. Kuhl, O. Hoegh-Guldberg, Fluorescent pigments in corals are photoprotective. Nature 2000, 408, 850.
| Fluorescent pigments in corals are photoprotective.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXhtlaisA%3D%3D&md5=31381ba64e719c9244cffa9bfd3a601bCAS | 11130722PubMed |
[58] A. R. J. Curson, J. D. Todd, M. J. Sullivan, A. W. B. Johnston, Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 2011, 9, 849.
| Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht12iurbE&md5=93732fa0bab3056569bbfa8e8bbadc36CAS |
[59] D. J. Schuller, C. R. Reisch, M. A. Moran, W. B. Whitman, W. N. Lanzilotta, Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagibacter ubique. Protein Sci. 2012, 21, 289.
| Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagibacter ubique.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XpslyqsQ%3D%3D&md5=8e56a89f533016c100058bcb0e208ee0CAS | 22162093PubMed |
[60] E. C. Howard, S. L. Sun, C. R. Reisch, D. A. del Valle, H. Burgmann, R. P. Kiene, M. A. Moran, Changes in dimethylsulfoniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl. Environ. Microbiol. 2011, 77, 524.
| Changes in dimethylsulfoniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXisVOqsb0%3D&md5=25f4455694980de315d2dc8f65dab6c8CAS | 21097583PubMed |
[61] C. R. Reisch, M. A. Moran, W. B. Whitman, Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 2008, 190, 8018.
| Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsV2lsLvK&md5=52fe46b2f71f5dd67a18921e4a0472bcCAS | 18849431PubMed |
[62] F. Rohwer, V. Seguritan, F. Azam, N. Knowlton, Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1.
| Diversity and distribution of coral-associated bacteria.Crossref | GoogleScholarGoogle Scholar |
[63] R. A. Littman, D. G. Bourne, B. L. Willis, Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol. Ecol. 2010, 19, 1978.
| Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXns1Gitr8%3D&md5=9272a0005aa06e0cc7804317cf4fc7c0CAS | 20529072PubMed |
[64] M. Vila-Costa, J. M. Rinta-Kanto, R. S. Poretsky, S. Sun, R. P. Kiene, M. A. Moran, Microbial controls on DMSP degradation and DMS formation in the Sargasso Sea. Biogeochemistry 2014, 120, 295.
| Microbial controls on DMSP degradation and DMS formation in the Sargasso Sea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXptVCrt7w%3D&md5=c06c5016ec78fe5de34cd9368bdb6ab8CAS |
[65] M. S. Savoca, G. A. Nevitt, Evidence that dimethyl sulfide facilitates a tritrophic mutualism between marine primary producers and top predators. Proc. Natl. Acad. Sci. USA 2014, 111, 4157.
| Evidence that dimethyl sulfide facilitates a tritrophic mutualism between marine primary producers and top predators.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXjtl2kt7w%3D&md5=935ded43fa889e5e490eb3ddb55b477eCAS | 24591607PubMed |