No evidence of rabbit haemorrhagic disease virus 2 infection in scavengers of wild rabbits in Australia
Ina L. Smith A * , Nina Huang A , Megan Pavy A , Alexander Gofton A , Omid Fahri A , Egi Kardia A B , Roslyn Mourant A , Sammi Chong A , Maria Jenckel A , Robyn N. Hall A C and Tanja Strive A
A * , Nina Huang A , Megan Pavy A , Alexander Gofton A , Omid Fahri A , Egi Kardia A B , Roslyn Mourant A , Sammi Chong A , Maria Jenckel A , Robyn N. Hall A C and Tanja Strive A
A
B Present address:
C Present address:
Abstract
Rabbit haemorrhagic disease virus 2 (RHDV2/b/GI.2) is the only representative of the genus Lagovirus that is known to fatally infect multiple lagomorph species. RHDV2 is the dominant lagovirus circulating in rabbits in Australia, where some lagoviruses are used for deliberate biological control of European rabbits, a major environmental and agricultural pest in this country. Evidence of exposure to lagoviruses has been reported for a range of species that feed on rabbits, and the reduced host specificity of RHDV2 compared with RHDV1 has occasionally raised concerns, especially in a biocontrol context.
We investigated evidence of exposure to RHDV2 in 99 individual feral foxes, cats, dogs and pigs and then we aimed to test these animals for evidence of a productive infection.
Sera were analysed for the presence of antibodies to RHDV2, and faeces and tissues for the presence of viral RNA. We made provisions for downstream analysis of liver tissues by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) and histopathology should they return a positive RT-qPCR result, to further investigate any evidence of productive virus infection. We also infected RHDV2 in hepatobiliary organoids derived from cats and foxes to test for possible infection.
We detected serum antibodies and viral RNA in faeces indicative of ingestion of RHDV2-infected rabbits, but found no evidence for productive infection with RHDV2. Furthermore, no RHDV2 replication was seen in hepatobiliary organoids derived from foxes and cats after in vitro infection with RHDV2.
RHDV2 does not infect scavengers of rabbits, such as foxes, dogs, cats and pigs.
This study has provided insights into the safety of this strain.
Keywords: hares, hepatobiliary organoids, lagomorphs, predators, rabbit haemorrhagic disease virus 2, rabbits, RHDV2, scavengers.
Introduction
Rabbit haemorrhagic disease virus (RHDV) has been used in Australia to control overabundant populations of European rabbits (Oryctolagus cuniculus), which are one of the most damaging environmental and agricultural pests in this country. RHDV belongs to the genus Lagovirus in the family Caliciviridae (Vinjé et al. 2019) and causes rabbit haemorrhagic disease (RHD), an infectious hepatitis with high case-fatality rates in the European rabbit. The virus first emerged in China in 1984 (Liu et al. 1984).
The first detected RHDV (RHDV1/GI.1; hereon referred to as RHDV1) was introduced as a biocontrol agent to Australia in 1995/96 and led to a widespread reduction and suppression of rabbit populations and impacts (Cooke and Fenner 2002). As part of the first release of RHDV as a rabbit biocontrol agent in the mid-1990s, scavengers feeding on rabbits were analysed to test whether these could be useful indicators for disease activity in sympatric rabbit populations, but also to alleviate public concerns of possible non-target effects. Serum antibodies against RHDV resulting from ingestion of large amounts of viral antigen were found in foxes (Vulpes vulpes) 4 months after reported outbreaks in rabbits, but no infectious virus was detected in the blood, and no viral antigen or RNA was detected in the liver (Philbey et al. 2005). This is in line with other studies reporting the presence of RHDV1 antibodies in scavengers (foxes, feral cats, stoats, ferrets, hedgehogs, hawks and gulls), but no evidence for productive infections (Leighton et al. 1995; Frölich et al. 1998; Zheng et al. 2003; Parkes et al. 2004; Philbey et al. 2005; Henning et al. 2006). Red foxes experimentally fed RHDV1-infected rabbit livers were found to produce a detectable immune response 7 days after ingestion, with antibodies still detectable at the end of the experiment, 6 months later. None of the red foxes developed clinical signs of disease (Leighton et al. 1995). RHDV1 was detected in faeces following experimental infection of dogs (Canis lupus familiaris) with virus-infected rabbit liver homogenates, and this faecal preparation was found to be infectious as disease was observed when fed to rabbits (Simón et al. 1994). Likewise, RHDV1 virus was detected by an antigen-capture ELISA in the faeces of cats 1–2 days after experimental ingestion of RHDV-infected livers (Zheng et al. 2003). In addition, caliciviruses have been found in faeces and gut contents of scavengers, suggesting a potential role in the dissemination of the virus in the environment, because virus can remain viable after passing through the gastrointestinal tract (Chiari et al. 2016; Di Profio et al. 2018). Disease was not identified in any of these studies. Similar findings have been reported for the related calicivirus, European brown hare syndrome virus (EBHSV), with detection of the virus in the intestinal contents from the caecum and rectum of red foxes by polymerase chain reaction (PCR) assay (Chiari et al. 2016) and from faecal swabs from 1 of 41 wolves (Canis lupus) and one of four foxes sampled, all from northern Italy (Di Profio et al. 2018).
In 2015, a new lagovirus was detected in Australia that was not deliberately released. This virus, RHDV2, first emerged in 2010 in France (Le Gall-Reculé et al. 2011) and has since spread worldwide (Rouco et al. 2018). In contrast to RHDV1, which fatally infects only rabbits of >4 weeks of age, rabbits of all ages are susceptible to fatal RHDV2 infection. RHDV2 also infects other members of the order Lagomorpha such as the European brown hare, Sardinian Cape hare (Lepus capensis mediterraneus) in Italy (Puggioni et al. 2013), the Italian hare (Lepus corsicanus) in Italy (Camarda et al. 2014), mountain hare (Lepus timidus) in Sweden (Neimanis et al. 2018), Irish hare (Lepus timidus hibernicus) in Ireland (Kennedy et al. 2021), scrub hares (Lepus saxatilis) in South Africa (Du Plessis 2022), and cottontails (Sylvilagus audubonii) and jackrabbits (Lepus alleni and Lepus californicus) in the United States of America (Asin et al. 2022), whereas RHDV1 is strictly species specific to European rabbits. In Australia, RHDV2 is now the dominant strain circulating in both wild and domestic rabbits (Peng et al. 2023) and has led to an average reduction of 60% in rabbit numbers since its incursion (Ramsey et al. 2023).
The broader host range of RHDV2 precipitated a series of studies investigating the possibility of host spillovers of RHDV2 into species outside of the lagomorph family. Some studies suggested spillover in the Mediterranean pine vole (Microtus duodecimcostatus) and the white-toothed shrew (Crocidura russula) (Calvete et al. 2019) on the basis of reverse transcription-PCR (RT-PCR) detection of low concentrations of viral RNA in liver samples, with the virus found to be infectious when laboratory rabbits were challenged. Several spillover cases in Eurasian badgers (Meles meles) were reported and showed RHDV2 antigen in the livers (Abade dos Santos et al. 2022); however, no further cases have been reported in this species since.
Host switches or spillover infections cannot be inferred solely from the presence of viral antigen in faeces or tissues, or antibodies to the virus, because both can be explained as a result of ingestion of large amounts of viral antigen or even environmental contamination, especially as lagoviruses replicate to very high titres in the host and are highly environmentally stable (Henning et al. 2005; Lavazza et al. 2023).
The arrival of RHDV2 in Australia again triggered speculations about the safety of viral biocontrol in general. RHDV2 RNA was detected in Australia in the faeces of scavengers such as the Tasmanian devil (Sarcophilus harrisii) (Chong et al. 2019) and in foxes in Sydney, New South Wales (Campbell et al. 2020), by using a metagenomic approach; however, no systematic studies were conducted to test for evidence of actual productive infections in hosts other than lagomorphs.
The aims of this study were two-fold. First, we hypothesised that, similar to RHDV1, scavengers of rabbits would show antibodies to RHDV2 and traces of viral RNA in faeces, which can serve as indicators of disease activity in sympatric rabbit or hare populations. Second, we aimed to test these scavengers for any evidence of productive viral infection, by collecting liver into paraformaldehyde for subsequent downstream analysis by histochemistry, immunohistochemistry or in situ hybridisation, to detect virus replication or newly synthesised viral antigens, or any histopathological changes where the matching liver sample had tested positive for viral RNA.
Furthermore, we utilised a recently described ex vivo culture system for lagoviruses to further investigate the species specificity of RHDV2 experimentally. Previous studies had shown that RHDV2 was unable to infect hepatobiliary organoids derived from cat and mouse livers (Kardia et al. 2023). We extended this work by inoculating hepatobiliary organoids derived from a fox liver with RHDV2 to observe for replication of the virus, so as to provide insights into host specificity.
Materials and methods
Fresh blood and tissue samples were opportunistically obtained from collaborations with private shooters, local land services (LLS groups), and park rangers in New South Wales (NSW), Australian Capital Territory (ACT), Queensland (Qld) and Western Australia (WA). Samples from individual scavengers or predators of rabbits (fox (Vulpes vulpes), cat (Felis catus), dog (Canis lupus familiaris) and pig (Sus scrofa domesticus)) were collected opportunistically as part of routine feral animal control operations, and therefore, ethical review and approval were waived by institutional review board for this study. Blood was collected and allowed to clot to acquire the serum. Liver was collected into RNAlater (20 mM EDTA, 25 mM sodium citrate and 70 g ammonium sulfate/100 mL pH 5.2) and paraformaldehyde (PFA), and faeces were collected in RNAlater, and an aliquot was stored frozen at −80°C. In total, 99 animals were sampled.
No liver samples had detectable RHDV2 viral RNA by qRT-PCR. The PFA-fixed livers from foxes (WM1, 74.2 and WM1) and cats (NC1, NC4) were submitted to the Elizabeth Macarthur Agricultural Institute (NSW Department of Primary Industries) for examination by histology to investigate whether the seropositive animals showed signs of disease.
RHDV2 serology and PCR
Antibodies to RHDV2 were analysed using an RHDV2-specific blocking ELISA, as previously described (Strive et al. 2020).
RNA was extracted from the liver and faecal samples preserved in RNAlater by using the Maxwell Simply RNA tissue extraction kit (Promega) on a Maxwell RSC extraction machine (Promega) according to the manufacturer’s instructions. Each extraction run included an uninfected rabbit liver control. A real-time RT-PCR targeting the VP60 gene for RHDV2 was performed to detect viral RNA (1 μL; Duarte et al. 2015) in a modified protocol by using the SensiFAST Probe No-ROX on-step kit (Bioline), by using primers at 0.4 μM of each primer (forward 5′-TGGAACTTGGCTTGAGTGTTGA-3′, reverse 5′-ACAAGCGTGCTTGTGGACGG-3′; Sigma) and 0.1 μM of probe (5′HEX – TGTCAGAACTTGTTGACATCCGCCC – 3′BHQ1; Sigma) in a final volume of 15 μL. Cycling was performed using a CFX-96 real-time system (Bio-Rad) in a 96-well optical plate with a reverse transcription at 45°C for 10 min, followed by an initial denaturation at 95°C for 2 min and then 40 cycles of 95°C for 5 s and extension at 63°C for 20 s. A fluorescence signal was acquired during the extension step. Negative template controls containing nuclease-free water instead of RNA were also included in each run.
Susceptibility of fox hepatobiliary organoids to RHDV2 infection
The fox liver organoids were established as detailed in Kardia et al. (2023). The fox liver sample was collected opportunistically during feral animal control activities. Monolayer cultures from fox, cat and rabbit liver organoids were prepared in eight-well chamber slides and inoculated in triplicate with RHDV2 with an infectious dose of 100 RID50 (50% rabbit infections dose) as described in Kardia et al. (2023), and heat-inactivated virus inoculum was used as a negative control. The experiment on all cultures was repeated to ensure reproducibility. Following overnight incubation, media was removed, and the cells were fixed in 4% paraformaldehyde for 15 min, followed by 10-min permeabilisation with 0.25% Triton X-100 in phosphate-buffered saline (PBS), and 60-min blocking in 5% bovine serum albumin in PBS at room temperature. The overnight incubation time was selected on the basis of previous work (Kardia et al. 2023), which showed this duration to be optimal for RHDV2 replication in vitro, allowing sufficient time for virus entry and initial stages of replication without compromising cell viability or monolayer integrity. Cells were incubated with a mouse monoclonal RHDV2-specific antibody against RNA-dependent RNA polymerase and 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) for DNA staining. Incubation for primary antibodies was overnight at 4°C and secondary antibodies (Alexa Fluor 488 labelled goat polyclonal secondary antibody to mouse IgG (ab150113, Abcam) for 1 h at room temperature. Cover slips were applied using Fluoroshield™ (Sigma-Aldrich) mounting medium. Images were taken using a Leica SP8 confocal microscope (Leica Microsystems) with Leica Application Suite X 3.5.1.18803. Two imaging tracks were used, one with 405 nm excitation for DAPI (signal collection at 420–440 nm) and another at 499 nm for Alexa Fluor™ 488 (signal collection at 510–540 nm), indicating the presence of RHDV2-RdRp and virus replication in permissive cells.
Results
In total, 99 feral animals, including 38 foxes (Vulpes vulpes), seven cats (Felis catus), eight dogs (Canis lupus familiaris) and 46 pigs (Sus scrofa domesticus) were sampled and analysed. Animals were obtained from Williams and Woodanilling in Western Australia (WA), Numerella, Murrumbateman, Mongarlowe, Bungendore, Braidwood, Mirrabooka and Williamsdale in New South Wales (NSW), Gudgenby in the Australian Capital Territory (ACT), and Wreck Beach and Rules Beach in Queensland (Qld).
All animals were healthy shot or trapped animals. We found evidence of exposure to RHDV2 in all species except dogs, by using serology; 2/13 WA foxes, 1/18 NSW foxes, 3/4 NSW cats and 1/46 NSW pigs had detectable antibodies against RHDV2 in the sera (Table 1). Two foxes had detectable viral RNA (expressed as cycle threshold (Ct) values) in their faeces, indicating that they had recently consumed RHDV2-infected rabbits. These foxes were collected during a period where there was a known outbreak of RHDV2 in the region. One fox and one cat had both detectable anti-RHDV2 antibodies in the sera and detectable RHDV2 RNA in their faeces. Overall, 13.2% (5/38) of foxes, 42.9% (3/7) of cats and 2.2% (1/46) of pigs had evidence of exposure to RHDV2.
| Sample | Collection date | State | Location | Species | RHDV2 TaqMan (Ct) | RHDV2 cELISA | ||
|---|---|---|---|---|---|---|---|---|
| RNAlater faeces | RNAlater liver | Serum titre | ||||||
| WM1 | 18 October 2020 | WA | Williams | Fox | 37.34 | Neg | 40 | |
| WD8 | 18 October 2020 | WA | Woodanilling | Fox | Neg | Neg | 40 | |
| WD10 | 18 October 2020 | WA | Woodanilling | Fox | 30.57 | Neg | <40 | |
| NF1 | 8 December 2020 | NSW | Numeralla | Fox | 32.54 | Neg | <40 | |
| 74.2 | 3 May 2022 | NSW | Bungendore | Fox | Neg | Neg | 80 | |
| NC1 | 5 December 2020 | NSW | Numeralla | Cat | Neg | Neg | 40 | |
| NC3 | 14 December 2020 | NSW | Numeralla | Cat | 28.74 | Neg | 40 | |
| NC4 | 16 December 2020 | NSW | Numeralla | Cat | Neg | Neg | 40 | |
| P4 | 22 October 2021 | NSW | Williamsdale | Pig | Neg | Neg | 40 | |
No animals had detectable viral RNA in the liver. No significant changes were observed following histological examination of the PFA-fixed liver from the seropositive animals.
Species specificity testing using organoids
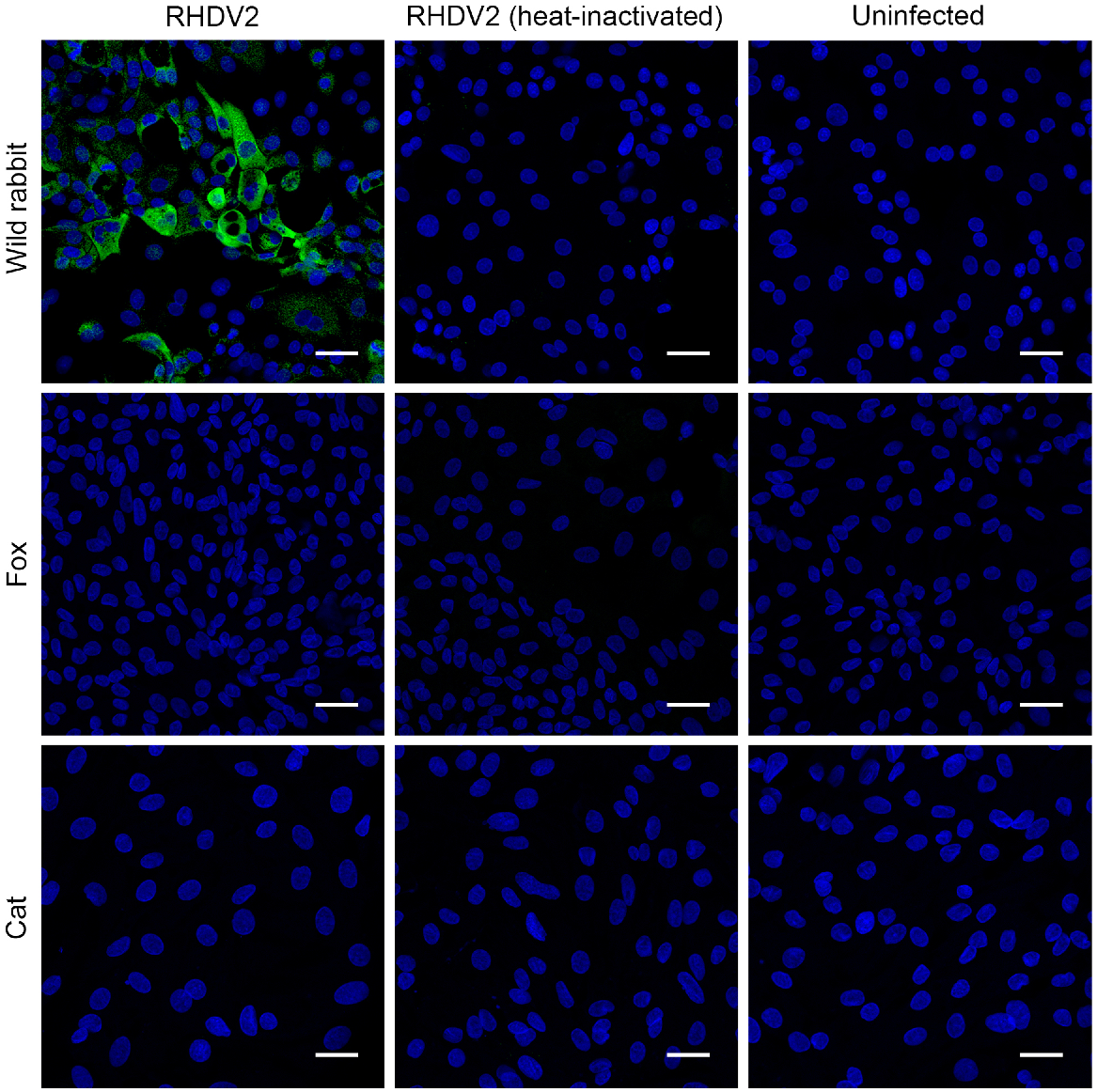
In line with previous reports, cat hepatobiliary organoids inoculated with RHDV2 did not show evidence of infection with RHDV2 (Kardia et al. 2023). Similarly, when RHDV2 was inoculated onto hepatobiliary organoids derived from a fox liver, no viral protein expression was detected in an immunofluorescence assay. For rabbit liver organoids, approximately 20–40% of cells showed positive staining for RHDV2-RdRp, confirming active replication in these permissive cells (Fig. 1).
Rabbit, fox, and cat hepatobiliary cells 24 h after inoculation. The cells, cultured on 8-chamber Nunc Lab-Tek II slides (Thermo Fisher Scientific), were inoculated with 100 RID50 of RHDV2 (left column). The control groups consisted of cells mock inoculated with culture medium (right column), and another group exposed to the same titre of heat-inactivated RHDV2 (middle column). Scale: 50 μm.
Discussion
Sampling was conducted on foxes, feral cats, dogs and pigs as part of pest control efforts targeting introduced species in Australia, aiming to assess their exposure to RHDV2-infected rabbits. All of the sampled animals were apparently healthy.
Similar to previously reported findings for animals that prey or scavenge potentially calicivirus-infected rabbits (Leighton et al. 1995; Frölich et al. 1998; Parkes et al. 2004; Philbey et al. 2005; Henning et al. 2006; Chiari et al. 2016; Di Profio et al. 2018; Campbell et al. 2020), we found evidence of consumption of RHDV-infected rabbits in five foxes, three cats and one pig.
A specific competitive ELISA was employed for the detection of RHDV2 antibodies in the serum of the animals. These antibodies may be generated following the ingestion of rabbits that have succumbed to RHDV2 infection, which contain very large amounts of viral particles (and therefore the antigenic VP60 capsid protein). In foxes, this serological response is known to develop 7 days after ingestion of RHDV1 and last for at least 6 months (Leighton et al. 1995). Antibodies against RHDV2 (7%) were detected in three foxes, three cats and one pig, with one fox and one cat also having detectable RHDV2 RNA in their faeces. An additional two foxes were seronegative for RHDV2, but had detectable RNA in their faeces by using a RHDV2-specific RT-qPCR assay, further indicating that these species consumed RHDV2-infected rabbits. This represented 4% of animals with detectable viral RNA in their faeces. The detection of RHDV2 in faeces of scavengers supports previous findings (Simón et al. 1994; Frölich et al. 1998; Chong et al. 2019; Campbell et al. 2020) that scavengers may act as mechanical vectors for RHDV, disseminating the virus in the environment, although we did not determine whether the viral RNA detected in faeces in this study was infectious.
In summary, 5 of the 38 foxes (13.2%), three of the seven cats (42.9%) and one of the pigs (2.2%) had evidence of exposure to RHDV2, as shown by the presence of serum antibodies to RHDV2, viral RNA in faeces, or both.
None of the animals had detectable viral RNA in their livers, nor were there any signs of viral infection observed in the matched PFA-fixed liver from the seropositive animals submitted for histological examination, which would have suggested a RHDV2 infection of these animals.
These results demonstrated that scavengers and predators of rabbits in Australia are being exposed to RHDV2, as observed by the seropositivity and presence of viral RNA in faeces. Similar to previous studies on RHDV1, scavengers can be an indicator of virus activity in sympatric lagomorph populations. This is not unexpected, given the amount of virus (107−1010 genome copies/mg of tissue) (Lavazza et al. 2023) that is contained in an infected rabbit carcass. There is currently no evidence that viral RNA is present in the liver (the target organ of RHDV) or replicating in the scavengers tested. Whereas no evidence of virus was detected in the tissues of the scavengers, this study outlined a sampling and testing protocol that incorporates histological analysis of liver samples from animals that had detectable antibodies in their sera and/or detectable RHDV2 RNA in the liver to enable a more robust analysis required to infer any suspected species jumps or viral spillover infections.
In line with previous studies, neither cat nor fox hepatobiliary organoids showed evidence of being permissive to infection with RHDV2 (Kardia et al. 2023). This provides further evidence that these species are not susceptible to infection with RHDV2 virus and highlights the value of the organoid platform for future studies on lagovirus species specificity testing.
In summary, this study sampled 99 animals that would naturally consume rabbits. The types of animals obtained were limited to those that were killed as part of feral control activities, and were sampled from a wide range across Australia, from central eastern Qld in the north, to Gudgenby in the ACT in the south, and as far west as Williams in WA. The study opportunistically sampled only a small number of feral cats and dogs. Despite the low sampling of these species, three of the four cats had evidence of having consumed RHDV2-infected rabbits. Additionally, this finding also suggests that rabbits constitute a portion of the diet of feral cats, another pest species in Australia.
Data availability
The data that support this study will be shared upon reasonable request to the corresponding author.
Conflicts of interest
The authors declare that they have no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the paper, or in the decision to publish the results.
Declaration of funding
This research was supported by Meat and Livestock Australia, Grant P.PSH.1059.
Author contributions
Conceptualisation, I.S., RH. M.J. and T.S.; methodology, N.H., M.P. A.G., O.F., E.K., R.M., S.C.; formal analysis, I.S; writing – original draft preparation, I.S.; writing – review and editing, all authors; supervision, I.S., T.S. and RH; project administration, I.S.; funding acquisition, T.S. All authors have read and agreed to the published version of the paper.
Acknowledgements
We thank Georgeanna Story from Upper Murrumbidgee Landcare; Richard and Alison Swain and Numeralla Landcare; Susan Campbell, Narelle Dybing and Heather Crawford from Murdoch University; Mick Davis, Dan Biddulph, and Nicky Webb from New South Wales Local Land Services and Darren Marshall from SQ Landscapes for assisting in providing samples from animal control activities for this study. We thank Pedro Pinczowski at the Elizabeth Macarthur Agriculture Institute for histological examination of the tissues. We thank Dr Phil Hands for training and assistance in collecting confocal microscopy results.
References
Abade dos Santos FA, Pinto A, Burgoyne T, Dalton KP, Carvalho CL, Ramilo DW, Carneiro C, Carvalho T, Peleteiro MC, Parra F, Duarte MD (2022) Spillover events of rabbit haemorrhagic disease virus 2 (recombinant GI.4P-GI.2) from Lagomorpha to Eurasian badger. Transboundary and Emerging Diseases 69(3), 1030-1045.
| Crossref | Google Scholar |
Asin J, Rejmanek D, Clifford DL, Mikolon AB, Henderson EE, Nyaoke AC, Macías-Rioseco M, Streitenberger N, Beingesser J, Woods LW, Lavazza A, Capucci L, Crossley B, Uzal FA (2022) Early circulation of rabbit haemorrhagic disease virus type 2 in domestic and wild lagomorphs in southern California, USA (2020–2021). Transboundary and Emerging Diseases 69(4), e394-e405.
| Crossref | Google Scholar | PubMed |
Calvete C, Mendoza M, Sarto MP, Bagüés MPJ, Luján L, Molín J, Calvo AJ, Monroy F, Calvo JH (2019) Detection of Rabbit Hemorrhagic Disease Virus GI.2/RHDV2/b in the Mediterranean Pine Vole (Microtus duodecimcostatus) and White-Toothed Shrew (Crocidura russula). Journal of Wildlife Diseases 55(2), 467-472.
| Crossref | Google Scholar | PubMed |
Camarda A, Pugliese N, Cavadini P, Circella E, Capucci L, Caroli A, Legretto M, Mallia E, Lavazza A (2014) Detection of the new emerging rabbit haemorrhagic disease type 2 virus (RHDV2) in Sicily from rabbit (Oryctolagus cuniculus) and Italian hare (Lepus corsicanus). Research in Veterinary Science 97(3), 642-645.
| Crossref | Google Scholar | PubMed |
Campbell SJ, Ashley W, Gil-Fernandez M, Newsome TM, Di Giallonardo F, Ortiz-Baez AS, Mahar JE, Towerton AL, Gillings M, Holmes EC, Carthey AJR, Geoghegan JL (2020) Red fox viromes in urban and rural landscapes. Virus Evolution 6(2), veaa065.
| Crossref | Google Scholar |
Chiari M, Molinari S, Cavadini P, Bertasi B, Zanoni M, Capucci L, Lavazza A (2016) Red foxes (Vulpes vulpes) feeding brown hares (Lepus europaeus) infected by European brown hare syndrome virus (EBHSv) might be involved in the spread of the virus. European Journal of Wildlife Research 62(6), 761-765.
| Crossref | Google Scholar |
Chong R, Shi M, Grueber CE, Holmes EC, Hogg CJ, Belov K, Barrs VR (2019) Fecal viral diversity of captive and wild tasmanian devils characterized using virion-enriched metagenomics and metatranscriptomics. Journal of Virology 93(11), e00205-e00219.
| Crossref | Google Scholar |
Cooke BD, Fenner F (2002) Rabbit haemorrhagic disease and the biological control of wild rabbits, Oryctolagus cuniculus, in Australia and New Zealand. Wildlife Research 29(6), 689-706.
| Crossref | Google Scholar |
Di Profio F, Melegari I, Sarchese V, Robetto S, Bermudez Sanchez S, Carella E, Orusa R, Cavadini P, Lavazza A, Marsilio F, Martella V, Di Martino B (2018) Potential role of wolf (Canis lupus) as passive carrier of European brown hare syndrome virus (EBHSV). Research in Veterinary Science 117, 81-84.
| Crossref | Google Scholar | PubMed |
Du Plessis J (2022) Update on the disease killing our rabbits: rabbit haemorrhagic disease virus. Available at https://ewt.org.za/update-on-the-disease-killing-our-rabbits/
Duarte MD, Carvalho CL, Barros SC, Henriques AM, Ramos F, Fagulha T, Luís T, Duarte EL, Fevereiro M (2015) A real time Taqman RT-PCR for the detection of rabbit hemorrhagic disease virus 2 (RHDV2). Journal of Virological Methods 219, 90-95.
| Crossref | Google Scholar | PubMed |
Frölich K, Kiima F, Dedek J (1998) Antibodies against rabbit hemorrhagic disease virus in free-ranging red foxes from Germany. Journal of Wildlife Diseases 34(3), 436-442.
| Google Scholar | PubMed |
Henning J, Meers J, Davies PR, Morris RS (2005) Survival of rabbit haemorrhagic disease virus (RHDV) in the environment. Epidemiology and Infection 133(4), 719-730.
| Crossref | Google Scholar | PubMed |
Henning J, Davies PR, Meers J (2006) Seropositivity to rabbit haemorrhagic disease virus in non-target mammals during periods of viral activity in a population of wild rabbits in New Zealand. Wildlife Research 33(4), 305-311.
| Crossref | Google Scholar |
Kardia E, Fakhri O, Pavy M, Mason H, Huang N, Smertina E, Jenckel M, Peng NYG, Estes MK, Strive T, Frese M, Smith I, Hall RN (2023) Hepatobiliary organoids derived from leporids support the replication of hepatotropic lagoviruses. Journal of General Virology 104(8), 001874.
| Crossref | Google Scholar |
Kennedy A, Britton L, Byrne AW, Byrne C, Casey M, Flynn O, Lozano JM, Marnell F, McElroy M, Reid N, Wilson M, FitzGerald W (2021) First detected case of rabbit Haemorrhagic disease virus 2 (RHDV2) in the Irish hare (Lepus timidus hibernicus). Irish Veterinary Journal 74(1), 25.
| Crossref | Google Scholar |
Lavazza A, Capucci L, Cavadini P (2023) Rabbit haemorrhagic disease. Manual of diagnostic tests and vaccines for terrestrial animals, Chapter 3.7.2, pp. 1–20. World Organisation for Animal Health. Available at https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/
Le Gall-Reculé G, Zwingelstein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, Decors A, Bertagnoli S, Guérin JL, Marchandeau S (2011) Detection of a new variant of rabbit haemorrhagic disease virus in France. Veterinary Record 168(5), 137-138.
| Crossref | Google Scholar |
Leighton FA, Artois M, Capucci L, Gavier-Widen D, Morisse J-P (1995) Antibody response to rabbit viral hemorrhagic disease virus in red foxes (Vulpes vulpes) consuming livers of infected rabbits (Oryctolagus cuniculus). Journal of Wildlife Diseases 31(4), 541-544.
| Crossref | Google Scholar | PubMed |
Liu SJ, Xue HP, Pu BQ, Qian NH (1984) A new viral disease in rabbits. Animal Husbandry and Veterinary Medicine 16(6), 253-255.
| Google Scholar |
Neimanis AS, Ahola H, Larsson Pettersson U, Lopes AM, Abrantes J, Zohari S, Esteves PJ, Gavier-Widén D (2018) Overcoming species barriers: an outbreak of Lagovirus europaeus GI.2/RHDV2 in an isolated population of mountain hares (Lepus timidus). BMC Veterinary Research 14(1), 367.
| Crossref | Google Scholar | PubMed |
Parkes JP, Heyward RP, Henning J, Motha MXJ (2004) Antibody responses to rabbit haemorrhagic disease virus in predators, scavengers, and hares in New Zealand during epidemics in sympatric rabbit populations. New Zealand Veterinary Journal 52(2), 85-89.
| Crossref | Google Scholar | PubMed |
Peng NYG, Hall RN, Huang N, West P, Cox TE, Mahar JE, Mason H, Campbell S, O’Connor T, Read AJ, Patel KK, Taggart PL, Smith IL, Strive T, Jenckel M (2023) Utilizing and citizen science for the surveillance of lagoviruses in Australia. Viruses 15(12), 2348.
| Crossref | Google Scholar |
Philbey AW, Kirkland PD, Saunders GR (2005) Assessment of antibodies to rabbit haemorrhagic disease virus in fox serum as an indicator of infection in sympatric rabbit populations. Australian Veterinary Journal 83(1–2), 97-100.
| Crossref | Google Scholar | PubMed |
Puggioni G, Cavadini P, Maestrale C, Scivoli R, Botti G, Ligios C, Le Gall-Reculé G, Lavazza A, Capucci L (2013) The new French 2010 rabbit hemorrhagic disease virus causes an RHD-like disease in the Sardinian Cape hare (Lepus capensis mediterraneus). Veterinary Research 44(1), 96.
| Crossref | Google Scholar | PubMed |
Ramsey DS, Patel KK, Campbell S, Hall RN, Taggart PL, Strive T (2023) Sustained impact of RHDV2 on wild rabbit populations across Australia eight years after its initial detection. Viruses 15(5), 1159.
| Crossref | Google Scholar |
Rouco C, Abrantes J, Serronha A, Lopes AM, Maio E, Magalhães MJ, Blanco E, Bárcena J, Esteves PJ, Santos N, Alves PC, Monterroso P (2018) Epidemiology of RHDV2 (Lagovirus europaeus/GI.2) in free-living wild European rabbits in Portugal. Transboundary and Emerging Diseases 65(2), e373-e382.
| Crossref | Google Scholar | PubMed |
Simón M, Muguruza R, Alonso J, Muzquiz J, Gironés O, Haffar A (1994) Recherche du virus de la maladie hémorragique virale du lapin (RHD) chez le renard et rôle des canidés domestiques dans la transmission de la maladie. Recueil de Medecine Veterinaire 170, 841-845.
| Google Scholar |
Strive T, Piper M, Huang N, Mourant R, Kovaliski J, Capucci L, Cox TE, Smith I (2020) Retrospective serological analysis reveals presence of the emerging lagovirus RHDV2 in Australia in wild rabbits at least five months prior to its first detection. Transboundary and Emerging Diseases 67(2), 822-833.
| Crossref | Google Scholar |
Vinjé J, Estes MK, Esteves P, Green KY, Katayama K, Knowles NJ, L’Homme Y, Martella V, Vennema H, White PA (2019) ICTV Virus Taxonomy Profile: Caliciviridae. Journal of General Virology 100(11), 1469-1470.
| Crossref | Google Scholar | PubMed |
Zheng T, Lu G, Napier AM, Lockyer SJ (2003) No virus replication in domestic cats fed with RHDV-infected rabbit livers. Veterinary Microbiology 95(1–2), 61-73.
| Crossref | Google Scholar | PubMed |


