Assessing freshwater fish biodiversity of Kumbe River, Papua (Indonesia) through environmental DNA metabarcoding
Arif Wibowo A * , Kurniawan Kurniawan A , Dwi Atminarso A B , Tri Heru Prihadi C , Lee J. Baumgartner B , Meaghan L. Rourke B D , Satoshi Nagai E , Nicolas Hubert F and Anti Vasemagi G H
A * , Kurniawan Kurniawan A , Dwi Atminarso A B , Tri Heru Prihadi C , Lee J. Baumgartner B , Meaghan L. Rourke B D , Satoshi Nagai E , Nicolas Hubert F and Anti Vasemagi G H
A Research Center for Conservation of Marine and Inland Water Resources, National Research and Innovation Agency, Jalan Raya Jakarta-Bogor Km. 48 Cibinong, Bogor, West Java 16911, Indonesia.
B Gulbali Institute for Agriculture, Water and Environment, Charles Sturt University, PO Box 789, Albury, NSW 2640, Australia.
C Research Center for Fisheries, National Research and Innovation Agency, Jalan Raya Jakarta-Bogor Km. 48 Cibinong, Bogor, West Java 16911, Indonesia.
D New South Wales Department of Primary Industries, Narrandera Fisheries Centre, Narrandera, NSW 2700, Australia.
E Japan Fisheries Research and Education Agency, Fisheries Resources Institute, Fisheries Stock Assessment Center, Yokohama, Japan.
F Université Montpellier (UMR) 5554 Institut des sciences de l’évolution de Montpellier (ISEM) (IRD, UM, CNRS, EPHE), Université de Montpellier, Place Eugène Bataillon, Montpellier cedex 05 34095, France.
G Department of Aquatic Resources, Institute of Freshwater Research, Swedish University of Agricultural Sciences, Drottningholm, Sweden.
H Estonian University of Life Sciences, Institute of Veterinary Medicine and Animal Sciences, Tartu, Estonia.
Pacific Conservation Biology 29(4) 340-350 https://doi.org/10.1071/PC21078
Submitted: 25 December 2021 Accepted: 29 July 2022 Published: 25 August 2022
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: The ability to accurately assess biodiversity is a critical first step towards effective conservation and management. However, assessment of biodiversity using conventional monitoring programs is often constrained by high cost and a lack of taxonomic expertise. Environmental DNA (eDNA) metabarcoding may be a useful tool to efficiently catalogue biodiversity in areas that cannot be easily assessed using other methods.
Aims: Here, we evaluated the potential of eDNA metabarcoding for assessing fish biodiversity and distribution in the Kumbe River, Papua Province, Indonesia.
Methods: We selected four sampling locations and collected seven eDNA samples from each location. We used eDNA metabarcoding of the Cytochrome-b gene to characterise the fish community.
Key results: A total of 23 species were detected, three of which comprised 92% of sequence reads detected: Melanotaenia goldiei (32%), Craterocephalus randi (31%), and the invasive tilapia Oreochromis niloticus (29%). Only five species that were previously detected using conventional methods were detected by metabarcoding: M. goldiei, Craterocephalus stercusmuscarum, O. niloticus, Neoarius graeffei, and Arius arius. We detected 18 species (70% native) that have never been recorded from the Kumbe River.
Conclusions: This work has demonstrated that fish biodiversity is substantially underestimated in the Kumbe River. Environmental DNA metabarcoding is a promising rapid, non-invasive and cost-effective method for assessing fish biodiversity in Papua.
Implications: The findings support future investment in eDNA metabarcoding to characterise the fish biodiversity in Papua. This will assist in allocating the limited resources for conservation and management to areas most at risk from anthropogenic impacts.
Keywords: biodiversity, eDNA, freshwater fish, Indonesia, metabarcoding, Papua, river, tropical.
Introduction
Defined by Walter Rosen in 1986 during the first American forum on biological diversity, biodiversity represents the variety and variability among living organisms and ecosystems where they occur (Pereira et al. 2013; Bartkowski et al. 2015). The challenge for biodiversity conservation is to understand when and how major perturbations such as climate change cause significant impact (Yachi and Loreau 1999; Mace et al. 2012). Thus, it is the responsibility of individual countries to preserve biodiversity for its environmental, productive, consumptive, social, ethical and aesthetic value (Pauchard 2017).
Indonesia is one of the most biodiverse countries, and its freshwater fish biodiversity is no exception. A total of 1200 native fish species have been reported across the archipelago (Kottelat and Whitten 1996; Hubert et al. 2015). The inventory of Indonesian freshwater species is ongoing, and recent studies have suggested that fish diversity may be underestimated (Kadarusman et al. 2012; Hubert et al. 2019; Sholihah et al. 2020, 2021a, 2021b). Thus, the incomplete understanding of fish biodiversity presents challenges for effective conservation and management. While traditional methods can be implemented to document biodiversity, these can be cost-prohibitive and time consuming. In addition, it also requires detailed expertise in fish taxonomy, which becomes limiting as the number of taxonomists declines. Optimising monitoring strategies will be critical to determine change in ecosystems; and monitoring over time is a crucial step toward sustainable use of resources (Xiao et al. 2016; Trebitz et al. 2017). This challenge is particularly pronounced in Indonesia, where aquatic biodiversity-related research output is not balanced with biodiversity complexity (Wibowo et al. 2018; Gustiano et al. 2021; Kurniawan et al. 2021) and understanding of the dynamics behind the origin and maintenance of freshwater biodiversity is still fragmentary (de Bruyn et al. 2013, 2014; Sholihah et al. 2021a, 2021b).
Threats to Indonesian aquatic biodiversity have escalated in recent decades through agricultural development (Cleary and DeVantier 2011; Imai et al. 2018; Austin et al. 2019), introduction of exotic species (Herder et al. 2012; Dahruddin et al. 2017), and pollution (Garg et al. 2018). As such, locally decreasing ichthyodiversity is of concern, and taxonomic knowledge gaps are challenging (Dahruddin et al. 2017; Hubert et al. 2019). Papua, the western most Indonesian province of the Island of New Guinea, contains 16% of known Indonesian freshwater fish diversity and 20% of endemic species, including all fish species within the genus Melanotaenia (Rainbowfish) (Hubert et al. 2015). New species or expanded distributions of known species are still being reported (Nugraha et al. 2015; Wibowo et al. 2017; Ditya et al. 2018). However, much of the fish biodiversity of Papua is still undiscovered (Kadarusman et al. 2012; Koh et al. 2013; Hubert et al. 2015).
An alternative approach to identify fish biodiversity is to collect DNA shed by the target species into the water. All aquatic organisms release DNA into the environment via mucus, urine, faeces, or dead tissue (Goldberg et al. 2016; Carraro et al. 2018), where it is then known as environmental DNA (eDNA). Water samples containing eDNA from the entire aquatic community (e.g. bacteria, algae, fish) can be filtered to extract eDNA and the target species can be identified using the relevant primer sets (Turner et al. 2015; Pont et al. 2018). Environmental DNA sampling can be used to detect a single species or multiple species (known as metabarcoding) (Taberlet et al. 2012; Rees et al. 2014; Pont et al. 2018). Although species-specific eDNA sampling may be more sensitive than metabarcoding – especially for rare species (Bylemans et al. 2019) – eDNA metabarcoding is a rapid, cost-effective, and non-invasive technique (Janosik and Johnston 2015; Seymour 2019; Sigsgaard et al. 2020). Environmental DNA has been successfully implemented in the surveillance of rare, invasive, or migratory fish species in various aquatic systems (Nathan et al. 2015; Sigsgaard et al. 2015; Simpfendorfer et al. 2016; Rice et al. 2018; Itakura et al. 2019; McElroy et al. 2020). However, eDNA metabarcoding has rarely been applied in Indonesia, with only three studies in freshwater environments. For instance, eDNA was successfully implemented in detection of invasive alligator gar Atractosteus spatula (Ulayya et al. 2020) and crayfish Cherax quadricarinatus (Djalil et al. 2018) in lakes of West Java. Assessing fish biodiversity using eDNA was carried out in the Maninjau lake of Sumatra, where 26 species were detected in the lake, including five native and 21 exotic fish species (Roesma et al. 2021).
The objective of this study was to evaluate the utility of eDNA metabarcoding to rapidly characterise the fish community of the Kumbe River in West Papua and assess its potential in a tropical context as a suitable alternative method to traditional sampling to characterise fish diversity.
Materials and methods
Sample collection and preservation
Sampling was undertaken at four locations in the Kumbe River, which is located in the southern part of Papua Island (Fig. 1). The Kumbe River is a meandering tropical river with a total length of 242 km and width ranging from 97 to 700 m (Anonymous 2010). The average annual rainfall of this area is 2160 mm and average annual temperature of 23°C (Peel et al. 2007). We collected seven 15 mL water samples (a total of 28 samples) from each of four sampling locations (Fig. 1). Immediately after collection, we added 1.5 mL of 3 M sodium acetate and 33 mL of absolute ethanol, mixed with the water sample and then stored it at room temperature in the field. Once in the laboratory, water samples were placed at −20°C for DNA preservation until DNA extraction.
DNA extraction and PCR amplification
Water samples were centrifuged for 35 min at 5500g at 24°C to precipitate DNA and suspended material, and supernatant was discarded (Valiere and Taberlet 2000). Precipitates were resuspended in 100 μL ultrapure water and DNA was extracted using the QIAamp Tissue Extraction Kit (Qiagen). We extracted DNA from each precipitate individually and DNA extraction was performed in a DNA laminar flow hood to avoid contamination. A negative control containing all reagents but no eDNA was included in each batch of extractions to monitor contamination. The concentration of extracted DNA ranged from 1 to 20 ng/μL. Polymerase chain reaction (PCR) amplification was performed using the primers 5′-TGCCAACGGAGCATCATTC-3′ and 5′-ATAAAGGTAGGAGCCGTAGT-3′, which amplify a 79 base pairs (bp) segment of the mitochondrial Cytochrome b (Cyt-b) (Ficetola et al. 2008). Each PCR reaction contained 2.5 μL of 1 × KAPA HiFi Hotstart ReadyMix (Kapa Biosystems), 0.5 μL of each primer (1 mM) and 5 μL of DNA extract; 4.0 μL PCR grade water was added up to a reaction volume of 12.5 μL. A negative control was included with each PCR batch to monitor the contamination of the reagents used. The PCR conditions were as follows: one cycle at 95°C for 5 min; 40 cycles at 95°C for 30 s, 50°C for 30 s, 72°C for 30 s; and one cycle at 72°C for 5 min. PCR products were visualised under UV light using a 2% agarose gel stained with Midori Green Advanced (Nippon Genetics Europe). PCR products were purified using the NucleoMag 96 PCR Kit (Macherey-Nagel).
Ion Torrent PGM preparation and reads
We pooled individually tagged DNA samples into two different sequencing libraries. Libraries were set up as described by Meyer and Kircher (2010) but modified as follows for Ion Torrent Personal Genome Machine (PGM) sequencing. The adapter mix was prepared using an adapter marked with a multiplex rating (MID). Adapters are designed to include individual MIDs and to match Ion Torrent-specific priming sites. The total molar concentration of the adapter mixture was 20 M for each adapter. We used a 100 bp long PCR product as the positive control DNA template. We carried out blunt-end reaction with half of the volume, i.e. we added 10 μL of blunt-end master mix to 25 μL of the sample.
The reaction was purified using solid phase reversible immobilisation (SPRI) beads with a template: bead ratio of 1:1.8. In the post-ligation purification steps, the template:bead ratio was 1:1. In order to verify adapter ligation success, the SPRI-purified MID-tagged positive control library and a subset of MID-tagged library aliquots were separated side by side with non-MID-tagged templates on 2% agarose gel stained with Midori Green Advanced electrophoresis (45 min, 95 V) and visualised using UV light. We measured DNA concentration of the samples using a Qubit Fluorometer (Life Technologies) and mixed amplicon libraries subsequently in equimolar ratios. The stock library was re-amplified in two different reactions as follows: 5 μL pooled library stock was added to a master mix comprising of 5 U of Herculase II polymerase (Agilent technologies, catalogue number 600677), 1 × Herculase II reaction buffer, 25 mM each deoxynucleoside triphosphate, 10 μM each primer, and added PCR grade water up to 50 μL. We designed the re-amplification primers based on the sequence of the IonTorrent-specific priming sites and thus generated millions of MID-tagged copies including binding sites necessary for subsequent sequencing with IonTorrent technology. The thermocycling profile included a 30 s denaturation at 98°C followed by 15 cycles consisting of a 20 s denaturation at 98°C, a 30 s annealing at 64°C, and a 30 s elongation at 72°C. Final elongation was conducted at 72°C for 5 min. To clean the re-amplified library, size-selection was conducted by separating the entire library using 2% Size-Select Agarose E- Gel and E-Gel Electrophoresis System (Life Technologies) according to the manufacturer’s instructions. The DNA concentration of amplified library pool was measured using a Qubit Fluorometer. The library pool stock was then diluted to a final concentration of 26 pmol. For template preparation, an 18 μL aliquot of the library dilution (approximately 2.8 × 108 molecules) was transferred into the sequencing reaction set-up. Emulsion PCR and Ion Torrent PGM sequencing were carried out on two 314 chips (Life Sciences, catalogue number 4462923) according to the manufacture’s protocol (Publication Part Number: 4471974 Rev. C).
The resulting reads were binned and renamed by MID, i.e. the original individual samples using the software Geneious Pro 6.1. Then the original-specific primers (Ficetola et al. 2008) were trimmed off, reads were trimmed for poor quality parts using a 0.05 error probability limit and reads shorter than 100 bp were excluded using Geneious Pro 6.1. Thus, only reads comprising both the Ion Torrent adapter with a MID and the original primer were passed on for further analysis. Subsequent analyses were performed employing supercomputers at the IT Centre for fisheries (Yokohama, Japan). Sequences were assigned to species using the BLASTN 2.2.25+ algorithm against the reference database (GenBank). We examined Basic Local Alignment Search Tool (BLAST) results showing a sequence similarity of at least 94% and we also considered species distribution when confirming species identity (Wibowo et al. 2017).
The analysis of BLAST result
The BLAST output was imported into the program MEGAN (Huson et al. 2007) to further summarise the results. We employed default parameters, except for following settings: Min support = 1, Min score = 100, Min complexity = 0. We utilised a built-in comparison tool in MEGAN for database comparison. We discarded all hits below 94% identity. We also discarded hits with e-value over 1e-20. We further analysed the hits one by one to see if there were reads matching more than one species in GenBank. In case of multiple hits, we (a) identified the hit with the longest alignment length, (b) identified the hit with a match to an adult specimen over a juvenile specimen (given the greater confidence in morphological identification of an adult specimen), and (c) reviewed the database entry for errors and omitted incorrect entries.
Results
A total of 629 976 raw reads were detected by sequencing of amplicons from 28 water samples and filtered to 297 438 reads. Thus, altogether 332 538 reads (52.79%) were discarded from further analysis after quality control. The numbers of filtered sequences obtained from Alfasera, Yakui, Sakor, and Inggun were 71 627, 89 014, 81 030 and 55 767 reads, respectively. BLAST queries resulted in sequence similarities between reads and reference sequences in Genbank ranging between 94 and 98% (Table 1). A total of 23 freshwater fish species were detected (Table 1).
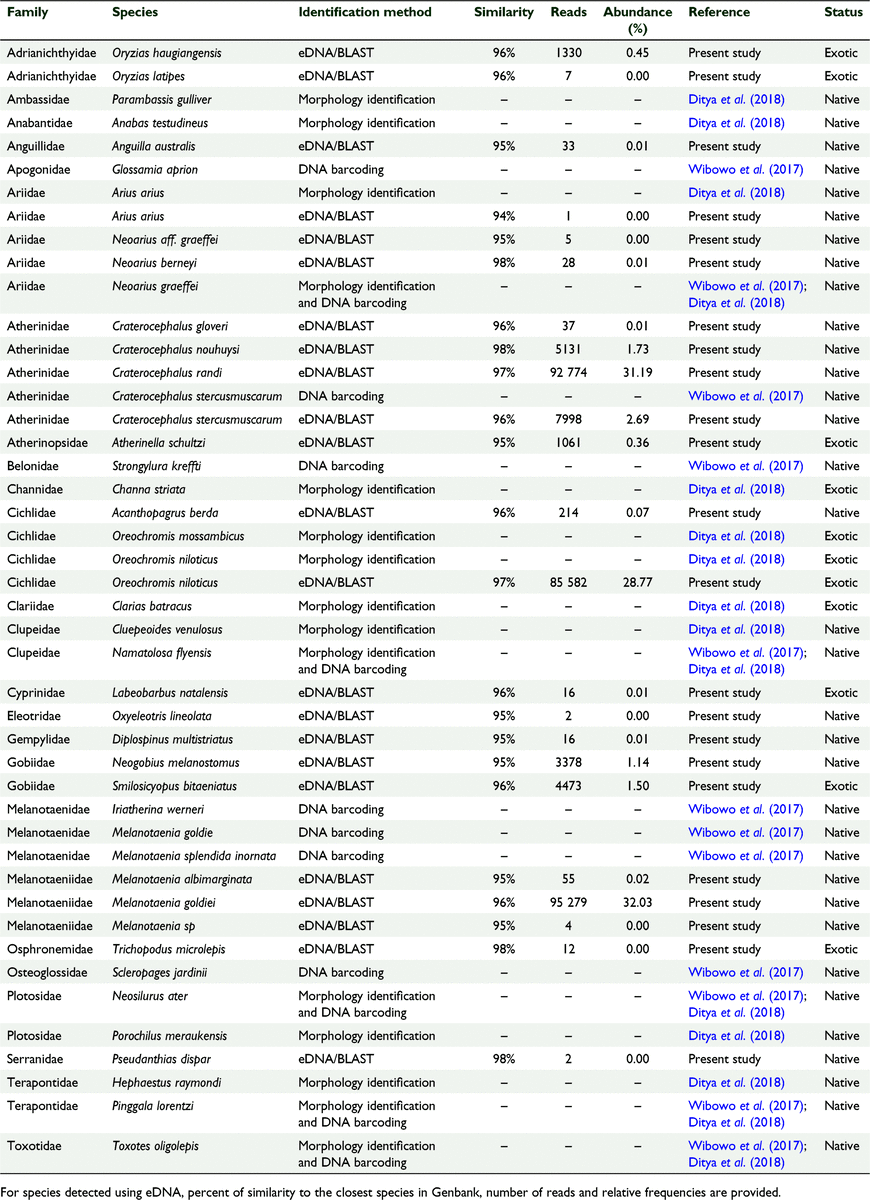
|
Species diversity was greatest at Inggun (21 species) and Yakui (21 species), followed by Sakor (19 species) and Alfasera (16 species) (Fig. 2a–d). The number of reads per species varied considerably among the seven sampling replicates within sites, with reads proportion ranging from 0.07 to 0.27 for Oreochromis niloticus, at Alfasera for instance (Fig. 2a). Some rarer species were not represented in all sampling replicates within sites including Trichopodus microlepis, Arius arius and Smilosicyopus bitaeniatus, which were not represented in replicates one and five at Yakui, and Melanotaenia goldiei, which was not represented in replicate six at Inggun.
|
|
Fourteen of the 23 species were detected at all sites (M. goldiei, O. niloticus, Craterocephalus randi, Craterocephalus stercusmuscarum, Craterocephalus nouhuysi, Craterocephalus gloveri, S. bitaeniatus, Neogobius melanostomus, Oryzias haugiangensis, Atherinella schultzi, Acanthopagrus berda, Diplospinus multistriatus, Anguilla australis and Labeobarbus natalensis), five species were detected at three sites (Melanotaenia albimarginata, M. sp, Neoarius graeffei, Neoarius berneyi and Oryzias latipes), two species were detected at in two sites (Oxyeleotris lineolata and Pseudanthias dispar) and two species were detected at one site (A. arius and T. microlepis) (Table 1, Fig. 3). A total of 16 species were native (M. goldiei, M. albimarginata, M. sp., C. randi, C. stercusmuscarum, C. nouhuysi, C. gloveri, N. melanostomus, N. berneyi, N. graeffei, A. berda, A. australis, D. multistriatus, O. lineolata, P. dispar and A. arius) and seven species were exotic (O. niloticus, S. bitaeniatus, O. haugiangensis, O. latipes, A. schultzi, L. natalensis and T. microlepis) (Table 1). If reads were ranked by abundance (Table 1), three species dominated all sites, consisting of M. goldiei (32.03% of the all reads), C. randi (31.19% of all reads), and O. niloticus (28.77% of reads). The 23 detected species belonged to 13 families and 16 genera. Atherinidae (35% of all sites; four species) was the most abundant family in terms of sequencing reads, followed by Melanotaeniidae (32% of all sites; three species) and Cichlidae (29% of all sites; two species). Sequences from these three families dominated all studied locations (Fig. 4a). These families mostly represented the genera Craterocephalus, Melanotaenia and Oreochromis (Fig. 4b).
|
|
A total of 23 species have previously been detected in the Kumbe River using conventional methods and DNA barcoding (Table 1). We detected five of these species (N. graeffei, M. goldie, C. stercusmuscarum, O. niloticus and A. arius) using metabarcoding, and an additional 18 species (69.56% native species) not previously recorded from the Kumbe River.
Discussion
Environmental DNA was successfully deployed to characterise freshwater fishes species richness and species occurrences at the Kumbe River. This represents one of the first eDNA metabarcoding assessments of fish biodiversity in Indonesia. Interestingly, N. graeffei or blue catfish was the dominant fish species (90%) in the Kumbe River based on monthly fishing over 2 years (Ditya et al. 2018). However, eDNA metabarcoding revealed only five N. graeffei reads from the whole dataset, while 91.9% of reads were mapped to only three species (M. goldiei, C. randi and O. niloticus). Thus, the lack of consensus between earlier studies employing traditional fishing methods (Wibowo et al. 2017; Ditya et al. 2018) and current eDNA analysis requires further investigation.
Our results showed that only five of the species we detected had been previously recorded (Wibowo et al. 2017; Ditya et al. 2018), and 18 represented new records, highlighting the limited knowledge of fish biodiversity in the Kumbe River. The detection of 18 new species is an important addition and highlights the critical role metabarcoding could have in rapidly describing fish biodiversity in tropical ecosystems. However, given that very low number of mapped reads (<50) for most of the species, the occurrence of these fishes in the River Kumbe needs to be validated by traditional approaches. Furthermore, it is possible that fish biodiversity in the Kumbe River is greater than reported here given many sequences were produced in this study that could not be identified due to gaps in the reference database. This demonstrates that it is critical that the reference databases be expanded to include more taxonomic groups to improve our understanding of tropical fish biodiversity (Schenekar et al. 2020) by taking every opportunity to collect fin clips from species where identification has been verified.
The inability of metabarcoding to detect 17 species previously recorded in the Kumbe River could be due to a range of factors. From a technical perspective, several reasons may account for this discrepancy: (1) some species may not have been present at the time of eDNA sampling (e.g. migratory species); (2) failure to detect a species using eDNA may be due to low abundance, for example, earlier studies have shown that eDNA metabarcoding may generate false negative results for rare species or lineages (Sato et al. 2017; Burian et al. 2021; Xiong et al. 2022); (3) the incomplete detection of species based on eDNA may be caused by insufficient volume of water filtered, filtration method, PCR inhibitors, primer bias, preferential amplification of abundant species, or an incomplete database of reference sequences (Deiner et al. 2017; Majaneva et al. 2018; Bessey et al. 2020; Schenekar et al. 2020; Rojahn et al. 2021).
We chose to use the Cyt-b gene fragment as it is commonly applied for fish metabarcoding studies. Some studies have demonstrated that Cyt-b may not perform as well as other regions, such as the 12S rRNA gene (Zhang et al. 2020; Shu et al. 2021). However, not all studies show consistent patterns and there may be distinct advantages and disadvantages with both markers (Hänfling et al. 2016). It is possible that the number of species detected may be maximised using multiple markers, which also enables the verification of taxa detected in each sample (Lecaudey et al. 2019). We therefore recommend that future metabarcoding studies in Indonesia evaluate the performance of multiple mitochondrial and nuclear markers to identify the most powerful targets for species identification.
From an ecological perspective, there are several limitations to characterising biodiversity based on eDNA metabarcoding. Environmental DNA production, degradation and transport are the three key factors that determine the quantity of eDNA in the water (Hansen et al. 2018). Thus several abiotic factors such as temperature, pH, dissolved oxygen and organic matter can degrade eDNA and reduce species detectability. In this study we did not account for any environmental factors, and thus detectability of some species may have been reduced. If eDNA metabarcoding was to become more widely applied in the tropical waters of Indonesia for conservation purposes – particularly for detection and quantification of specific species – we recommend further research into the role that abiotic and biotic factors play in eDNA production and degradation (Stewart 2019). In addition, false-positives detections may be caused by misidentification of taxa or contamination during field sampling or in the laboratory (Sato et al. 2017; Fujii et al. 2019). Thus, more comparative studies at species-rich regions are needed to evaluate when eDNA metabarcoding estimates of species richness are on par with, or potentially better than traditional fisheries methods (Ficetola et al. 2015; Olds et al. 2016; Cilleros et al. 2019). Nevertheless, despite the limitations, we have demonstrated that eDNA metabarcoding is a promising method to assess biodiversity in Indonesia. We recommend that eDNA metabarcoding be deployed concurrently with traditional methods for optimal results while methods are being optimised and reference databases are being improved.
Fish biomonitoring using eDNA has been implemented in the detection of invasive species and assessing their distribution (Nathan et al. 2015; Dunker et al. 2016). The invasive Nile tilapia O. niloticus was detected at all sites, which is concerning given it is implicated in extirpations of native fish species (Ogutu-Ohwayo 1990; Ligtvoet et al. 1991). The Nile tilapia is an important source of animal protein across its native range, and as such, it has been widely introduced in Papua for aquaculture purposes. Environmental DNA metabarcoding in Indonesian waterways may be a rapid and cost-effective method to track new introductions or expansion of Nile tilapia.
The Kumbe River drains into the Arafura Sea and is expected to contain a diverse fish community comprised of potamodromous species as well as species that require access to both freshwater and seawater to complete their lifecycle. Given the Kumbe River is a pristine aquatic habitat where land use has had minimal negative impacts on the environment (Wibowo et al. 2017; Lasmana et al. 2018), eDNA metabarcoding could be used to determine if migratory species are able to access upstream habitat as expected (Duda et al. 2021). In addition, eDNA metabarcoding could also be used to monitor spawning patterns of migratory fish species (either potamodromous or diadromous) (Yamanaka and Minamoto 2016; Thalinger et al. 2019). For example, if migratory species diversity is similar at upstream and downstream locations, this indicates good habitat connectivity between studied locations (Yamanaka and Minamoto 2016; Carraro et al. 2018). We detected A. australis, known as catadromous species, at the most upstream site (Alfasera). This likely suggests that the lower reaches of the Kumbe River, where this study was conducted, have sufficient connectivity to allow fish passage upstream. However, as this species is able to climb over migration barriers (Jellyman et al. 2017), A. australis presence upstream may not indicate that there is clear passage upstream for other species with lower upstream migration capabilities.
Conclusion
Environmental DNA metabarcoding shows great promise as a novel tool for fish biomonitoring programs in tropical freshwater habitats. It is sensitive, non-invasive and cost-effective as well as enabling a rapid assessment for detection of invasive, rare or endangered species. It provides an alternative approach when traditional sampling is time consuming and requires taxonomic expertise. However, our results highlight that eDNA metabarcoding cannot be routinely applied without further research into sampling design, marker choice, seasonal data and an improved reference databases to reduce false negative detections. Despite this, eDNA metabarcoding will play an important role in the future characterisation of the fish community in tropical river systems.
Data availability
The data used in this manuscript are organised by the first author (Arif Wibowo). Access to this data can be negotiated with him.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This study was supported financially by the Research Institute for Inland Fisheries and Extensions (RIIFE), Division of Genetics, Department of Biology, Turku University, the Erasmus Mundus Research Fellowship Programme, Academy of Finland, and the Estonian Ministry of Education and Research (Institutional Research Funding Project IUT8–2).
Acknowledgements
We thank Research Institute for Inland Fisheries and Extensions for all support including staff and budget through DIPA 2017–2019. We are also grateful to officers from local fisheries districts in West Papua for sampling collection and fruitful discussion and finally all the reviewers for improving the manuscript.
References
Anonymous (2010) Inventarisation of river in Papua, Indonesia in 2010. Available at https://lingkunganhidup.papua.go.id/gi/fckimage/file/SLHD/Tabel%20SD-11.pdfAustin, KG, Schwantes, A, Gu, Y, and Kasibhatla, PS (2019). What causes deforestation in Indonesia? Environmental Research Letters 14, 024007.
| What causes deforestation in Indonesia?Crossref | GoogleScholarGoogle Scholar |
Bartkowski, B, Lienhoop, N, and Hansjürgens, B (2015). Capturing the complexity of biodiversity: a critical review of economic valuation studies of biological diversity. Ecological Economics 113, 1–14.
| Capturing the complexity of biodiversity: a critical review of economic valuation studies of biological diversity.Crossref | GoogleScholarGoogle Scholar |
Bessey, C, Jarman, SN, Berry, O, Olsen, YS, Bunce, M, Simpson, T, Power, M, McLaughlin, J, Edgar, GJ, and Keesing, J (2020). Maximizing fish detection with eDNA metabarcoding. Environmental DNA 2, 493–504.
| Maximizing fish detection with eDNA metabarcoding.Crossref | GoogleScholarGoogle Scholar |
Burian, A, Mauvisseau, Q, Bulling, M, Domisch, S, Qian, S, and Sweet, M (2021). Improving the reliability of eDNA data interpretation. Molecular Ecology Resources 21, 1422–1433.
| Improving the reliability of eDNA data interpretation.Crossref | GoogleScholarGoogle Scholar |
Bylemans, J, Gleeson, DM, Duncan, RP, Hardy, CM, and Furlan, EM (2019). A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes. Environmental DNA 1, 402–414.
| A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes.Crossref | GoogleScholarGoogle Scholar |
Carraro, L, Hartikainen, H, Jokela, J, Bertuzzo, E, and Rinaldo, A (2018). Estimating species distribution and abundance in river networks using environmental DNA. Proceedings of the National Academy of Sciences 115, 11724–11729.
| Estimating species distribution and abundance in river networks using environmental DNA.Crossref | GoogleScholarGoogle Scholar |
Cilleros, K, Valentini, A, Allard, L, Dejean, T, Etienne, R, Grenouillet, G, Iribar, A, Taberlet, P, Vigouroux, R, and Brosse, S (2019). Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): a test with Guianese freshwater fishes. Molecular Ecology Resources 19, 27–46.
| Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): a test with Guianese freshwater fishes.Crossref | GoogleScholarGoogle Scholar |
Cleary DFR, DeVantier L (2011) Indonesia: threats to the country’s biodiversity. In ‘Encyclopedia of environmental health’. (Ed. JO Nriagu) pp. 187–197. (Elsevier)
Dahruddin, H, Hutama, A, Busson, F, Sauri, S, Hanner, R, Keith, P, Hadiaty, R, and Hubert, N (2017). Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species. Molecular Ecology Resources 17, 288–299.
| Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species.Crossref | GoogleScholarGoogle Scholar |
de Bruyn, M, Rüber, L, Nylinder, S, Stelbrink, B, Lovejoy, NR, Lavoué, S, Tan, HH, Nugroho, E, Wowor, D, Ng, PKL, Siti Azizah, MN, Von Rintelen, T, Hall, R, and Carvalho, GR (2013). Paleo-drainage basin connectivity predicts evolutionary relationships across three Southeast Asian biodiversity hotspots. Systematic Biology 62, 398–410.
| Paleo-drainage basin connectivity predicts evolutionary relationships across three Southeast Asian biodiversity hotspots.Crossref | GoogleScholarGoogle Scholar |
de Bruyn, M, Stelbrink, B, Morley, RJ, Hall, R, Carvalho, GR, Cannon, CH, van den Bergh, G, Meijaard, E, Metcalfe, I, Boitani, L, Maiorano, L, Shoup, R, and von Rintelen, T (2014). Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Systematic Biology 63, 879–901.
| Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity.Crossref | GoogleScholarGoogle Scholar |
Deiner, K, Bik, HM, Mächler, E, Seymour, M, Lacoursière-Roussel, A, Altermatt, F, Creer, S, Bista, I, Lodge, DM, de Vere, N, Pfrender, ME, and Bernatchez, L (2017). Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Molecular Ecology 26, 5872–5895.
| Environmental DNA metabarcoding: transforming how we survey animal and plant communities.Crossref | GoogleScholarGoogle Scholar |
Ditya, YC, Wibowo, A, and Husnah, H (2018). Spatial and temporal distribution of fish in the floodplain of Kumbe River. Papua. Indonesian Fisheries Research Journal 24, 69–81.
| Spatial and temporal distribution of fish in the floodplain of Kumbe River. Papua.Crossref | GoogleScholarGoogle Scholar |
Djalil, VN, Farajallah, A, and Wardiyatno, Y (2018). Application of environmental DNA (eDNA) for detection of Cherax quadricarinatus (Von Martens 1868) using water sample. Jurnal Biologi Tropis 18, 134–140.
| Application of environmental DNA (eDNA) for detection of Cherax quadricarinatus (Von Martens 1868) using water sample.Crossref | GoogleScholarGoogle Scholar |
Duda, JJ, Hoy, MS, Chase, DM, Pess, GR, Brenkman, SJ, McHenry, MM, and Ostberg, CO (2021). Environmental DNA is an effective tool to track recolonizing migratory fish following large-scale dam removal. Environmental DNA 3, 121–141.
| Environmental DNA is an effective tool to track recolonizing migratory fish following large-scale dam removal.Crossref | GoogleScholarGoogle Scholar |
Dunker, KJ, Sepulveda, AJ, Massengill, RL, Olsen, JB, Russ, OL, Wenburg, JK, and Antonovich, A (2016). Potential of environmental DNA to evaluate Northern Pike (Esox lucius) eradication efforts: an experimental test and case study. PLoS ONE 11, e0162277.
| Potential of environmental DNA to evaluate Northern Pike (Esox lucius) eradication efforts: an experimental test and case study.Crossref | GoogleScholarGoogle Scholar |
Ficetola, GF, Miaud, C, Pompanon, F, and Taberlet, P (2008). Species detection using environmental DNA from water samples. Biology Letters 4, 423–425.
| Species detection using environmental DNA from water samples.Crossref | GoogleScholarGoogle Scholar |
Ficetola, GF, Pansu, J, Bonin, A, Coissac, E, Giguet-Covex, C, De Barba, M, Gielly, L, Lopes, CM, Boyer, F, Pompanon, F, Raye, G, and Taberlet, P (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Molecular Ecology Resources 15, 543–556.
| Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data.Crossref | GoogleScholarGoogle Scholar |
Fujii, K, Doi, H, Matsuoka, S, Nagano, M, Sato, H, and Yamanaka, H (2019). Environmental DNA metabarcoding for fish community analysis in backwater lakes: a comparison of capture methods. PLoS ONE 14, e0210357.
| Environmental DNA metabarcoding for fish community analysis in backwater lakes: a comparison of capture methods.Crossref | GoogleScholarGoogle Scholar |
Garg, T, Hamilton, SE, Hochard, JP, Kresch, EP, and Talbot, J (2018). (Not so) gently down the stream: river pollution and health in Indonesia. Journal of Environmental Economics and Management 92, 35–53.
| (Not so) gently down the stream: river pollution and health in Indonesia.Crossref | GoogleScholarGoogle Scholar |
Goldberg, CS, Turner, CR, Deiner, K, Klymus, KE, Thomsen, PF, Murphy, MA, Spear, SF, McKee, A, Oyler-McCance, SJ, Cornman, RS, Laramie, MB, Mahon, AR, Lance, RF, Pilliod, DS, Strickler, KM, Waits, LP, Fremier, AK, Takahara, T, Herder, JE, Taberlet, P, and Gilbert, M (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution 7, 1299–1307.
| Critical considerations for the application of environmental DNA methods to detect aquatic species.Crossref | GoogleScholarGoogle Scholar |
Gustiano, R, Kurniawan, K, and Haryono, H (2021). Optimizing the utilization of genetic resources of Indonesian native freshwater fish. Asian Journal of Conservation Biology 10, 189–196.
| Optimizing the utilization of genetic resources of Indonesian native freshwater fish.Crossref | GoogleScholarGoogle Scholar |
Hänfling, B, Lawson Handley, L, Read, DS, Hahn, C, Li, J, Nichols, P, Blackman, RC, Oliver, A, and Winfield, IJ (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Molecular Ecology 25, 3101–3119.
| Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods.Crossref | GoogleScholarGoogle Scholar |
Hansen, BK, Bekkevold, D, Clausen, LW, and Nielsen, EE (2018). The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries. Fish and Fisheries 19, 751–768.
| The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries.Crossref | GoogleScholarGoogle Scholar |
Herder, F, Schliewen, UK, Geiger, MF, Hadiaty, RK, Gray, SM, McKinnon, JS, Walter, RP, and Pfaender, J (2012). Alien invasion in Wallace’s dreamponds: records of the hybridogenic “flowerhorn” cichlid in Lake Matano, with an annotated checklist of fish species introduced to the Malili Lakes system in Sulawesi. Aquatic Invasions 7, 521–535.
| Alien invasion in Wallace’s dreamponds: records of the hybridogenic “flowerhorn” cichlid in Lake Matano, with an annotated checklist of fish species introduced to the Malili Lakes system in Sulawesi.Crossref | GoogleScholarGoogle Scholar |
Hubert, N, Kadarusman, , Wibowo, A, Busson, F, Caruso, D, Sulandari, S, Nafiqoh, N, Pouyaud, L, Rüber, L, Avarre, J-C, Herder, F, Hanner, R, Keith, P, and Hadiaty, RK (2015). DNA barcoding Indonesian freshwater fishes: challenges and prospects. DNA Barcodes 3, 144–169.
| DNA barcoding Indonesian freshwater fishes: challenges and prospects.Crossref | GoogleScholarGoogle Scholar |
Hubert, N, Lumbantobing, D, Sholihah, A, Dahruddin, H, Delrieu-Trottin, E, Busson, F, Sauri, S, Hadiaty, R, and Keith, P (2019). Revisiting species boundaries and distribution ranges of Nemacheilus spp. (Cypriniformes: Nemacheilidae) and Rasbora spp. (Cypriniformes: Cyprinidae) in Java, Bali and Lombok through DNA barcodes: implications for conservation in a biodiversity hotspot. Conservation Genetics 20, 517–529.
| Revisiting species boundaries and distribution ranges of Nemacheilus spp. (Cypriniformes: Nemacheilidae) and Rasbora spp. (Cypriniformes: Cyprinidae) in Java, Bali and Lombok through DNA barcodes: implications for conservation in a biodiversity hotspot.Crossref | GoogleScholarGoogle Scholar |
Huson, DH, Auch, AF, Qi, J, and Schuster, SC (2007). MEGAN analysis of metagenomic data. Genome Research 17, 377–386.
| MEGAN analysis of metagenomic data.Crossref | GoogleScholarGoogle Scholar |
Imai, N, Furukawa, T, Tsujino, R, Kitamura, S, and Yumoto, T (2018). Factors affecting forest area change in Southeast Asia during 1980–2010. PLoS ONE 13, e0197391.
| Factors affecting forest area change in Southeast Asia during 1980–2010.Crossref | GoogleScholarGoogle Scholar |
Itakura, H, Wakiya, R, Yamamoto, S, Kaifu, K, Sato, T, and Minamoto, T (2019). Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scale. Aquatic Conservation: Marine and Freshwater Ecosystems 29, 361–373.
| Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scale.Crossref | GoogleScholarGoogle Scholar |
Janosik, AM, and Johnston, CE (2015). Environmental DNA as an effective tool for detection of imperiled fishes. Environmental Biology of Fishes 98, 1889–1893.
| Environmental DNA as an effective tool for detection of imperiled fishes.Crossref | GoogleScholarGoogle Scholar |
Jellyman, PG, Bauld, JT, and Crow, SK (2017). The effect of ramp slope and surface type on the climbing success of shortfin eel (Anguilla australis) elvers. Marine and Freshwater Research 68, 1317–1324.
| The effect of ramp slope and surface type on the climbing success of shortfin eel (Anguilla australis) elvers.Crossref | GoogleScholarGoogle Scholar |
Kadarusman, , Hubert, N, Hadiaty, RK, Sudarto, , Paradis, E, and Pouyaud, L (2012). Cryptic diversity in Indo-Australian rainbowfishes revealed by DNA barcoding: implications for conservation in a biodiversity hotspot candidate. PLoS ONE 7, e40627.
| Cryptic diversity in Indo-Australian rainbowfishes revealed by DNA barcoding: implications for conservation in a biodiversity hotspot candidate.Crossref | GoogleScholarGoogle Scholar |
Koh LP, Kettle CJ, Sheil D, Lee TM, Giam X, Gibson L, Clements GR (2013) Biodiversity state and trends in Southeast Asia. In ‘Encyclopedia of biodiversity’. (Ed. SA Levin) pp. 509–527. (Academic Press)
Kottelat M, Whitten T (1996) Freshwater biodiversity in Asia with special reference to fish. World Bank Technical paper. (World Bank)
Kurniawan, K, Gustiano, R, Kusmini, II, and Prakoso, VA (2021). Genetic resources preservation and utilization of Indonesian native freshwater fish consumption. Ecology, Environment and Conservation 27, 227–233.
Lasmana, Y, Simanungkalit, P, Gifariyono, M, Sotyadarpita, G, and Triadi, LB (2018). Potential of tidal lowland for irrigation development in Merauke Regency using hydrodynamic modelling 1D2D. Jurnal Teknik Hidraulik 9, 17–32.
| Potential of tidal lowland for irrigation development in Merauke Regency using hydrodynamic modelling 1D2D.Crossref | GoogleScholarGoogle Scholar |
Lecaudey, LA, Schletterer, M, Kuzovlev, VV, Hahn, C, and Weiss, SJ (2019). Fish diversity assessment in the headwaters of the Volga River using environmental DNA metabarcoding. Aquatic Conservation: Marine and Freshwater Ecosystems 29, 1785–1800.
| Fish diversity assessment in the headwaters of the Volga River using environmental DNA metabarcoding.Crossref | GoogleScholarGoogle Scholar |
Ligtvoet, W, Witte, F, Goldschmidt, T, van Oijen, MJP, Wanink, JH, and Goudswaard, PC (1991). Species extinction and concomitant ecological changes in Lake Victoria. Netherlands Journal of Zoology 42, 214–232.
| Species extinction and concomitant ecological changes in Lake Victoria.Crossref | GoogleScholarGoogle Scholar |
Mace, GM, Norris, K, and Fitter, AH (2012). Biodiversity and ecosystem services: a multilayered relationship. Trends in Ecology & Evolution 27, 19–26.
| Biodiversity and ecosystem services: a multilayered relationship.Crossref | GoogleScholarGoogle Scholar |
Majaneva, M, Diserud, OH, Eagle, SHC, Bostrom, E, Hajibabaei, M, and Ekrem, T (2018). Environmental DNA filtration techniques affect recovered biodiversity. Scientific Reports 8, 4682.
| Environmental DNA filtration techniques affect recovered biodiversity.Crossref | GoogleScholarGoogle Scholar |
McElroy, ME, Dressler, TL, Titcomb, GC, Wilson, EA, Deiner, K, Dudley, TL, Eliason, EJ, Evans, NT, Gaines, SD, Lafferty, KD, Lamberti, GA, Li, Y, Lodge, DM, Love, MS, Mahon, AR, Pfrender, ME, Renshaw, MA, Selkoe, KA, and Jerde, CL (2020). Calibrating environmental DNA metabarcoding to conventional surveys for measuring fish species richness. Frontiers in Ecology and Evolution 8, 276.
| Calibrating environmental DNA metabarcoding to conventional surveys for measuring fish species richness.Crossref | GoogleScholarGoogle Scholar |
Meyer, M, and Kircher, M (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbour Protocols 6, 1–10.
Nathan, LR, Jerde, CL, Budny, ML, and Mahon, AR (2015). The use of environmental DNA in invasive species surveillance of the Great Lakes commercial bait trade. Conservation Biology 29, 430–439.
| The use of environmental DNA in invasive species surveillance of the Great Lakes commercial bait trade.Crossref | GoogleScholarGoogle Scholar |
Nugraha, MFI, Pouyaud, L, Carman, O, Widyastuti, U, Junior, MZ, Kadarusman, , and Avarre, J-C (2015). Genetic diversity of Boeseman’s rainbowfish (Melanotaenia boesemani) reared in Indonesian farms compared to endangered natural populations. Tropical Conservation Science 8, 796–812.
| Genetic diversity of Boeseman’s rainbowfish (Melanotaenia boesemani) reared in Indonesian farms compared to endangered natural populations.Crossref | GoogleScholarGoogle Scholar |
Ogutu-Ohwayo, R (1990). The decline of native fishes of Lake Victoria and Kyoga (East Africa) and the impact of the introduced species, other environmental characteristics on amphibian distribution and abundance in mountain lakes of Northern Spain. Animal Conservation 9, 171–178.
Olds, BP, Jerde, CL, Renshaw, MA, Li, Y, Evans, NT, Turner, CR, Deiner, K, Mahon, AR, Brueseke, MA, Shirey, PD, Pfrender, ME, Lodge, DM, and Lamberti, GA (2016). Estimating species richness using environmental DNA. Ecology and Evolution 6, 4214–4226.
| Estimating species richness using environmental DNA.Crossref | GoogleScholarGoogle Scholar |
Pauchard, N (2017). Access and benefit sharing under the convention on biological diversity and its protocol: what can some numbers tell us about the effectiveness of the regulatory regime? Resources 6, 11.
| Access and benefit sharing under the convention on biological diversity and its protocol: what can some numbers tell us about the effectiveness of the regulatory regime?Crossref | GoogleScholarGoogle Scholar |
Peel, MC, Finlayson, BL, and McMahon, TA (2007). Updated world map of the Köppen-Geiger climate classification. Hydrology and Earth System Sciences 11, 1633–1644.
| Updated world map of the Köppen-Geiger climate classification.Crossref | GoogleScholarGoogle Scholar |
Pereira, HM, Ferrier, S, Walters, M, Geller, GN, Jongman, RHG, Scholes, RJ, Bruford, MW, Brummitt, N, Butchart, SHM, Cardoso, AC, Coops, NC, Dulloo, E, Faith, DP, Freyhof, J, Gregory, RD, Heip, C, Höft, R, Hurtt, G, Jetz, W, Karp, DS, McGeoch, MA, Obura, D, Onoda, Y, Pettorelli, N, Reyers, B, Sayre, R, Scharlemann, JPW, Stuart, SN, Turak, E, Walpole, M, and Wegmann, M (2013). Essential biodiversity variables. Science 339, 277–278.
| Essential biodiversity variables.Crossref | GoogleScholarGoogle Scholar |
Pont, D, Rocle, M, Valentini, A, Civade, R, Jean, P, Maire, A, Roset, N, Schabuss, M, Zornig, H, and Dejean, T (2018). Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Scientific Reports 8, 10361.
| Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation.Crossref | GoogleScholarGoogle Scholar |
Rees, HC, Maddison, BC, Middleditch, DJ, Patmore, JRM, and Gough, KC (2014). REVIEW: the detection of aquatic animal species using environmental DNA – a review of eDNA as a survey tool in ecology. Journal of Applied Ecology 51, 1450–1459.
| REVIEW: the detection of aquatic animal species using environmental DNA – a review of eDNA as a survey tool in ecology.Crossref | GoogleScholarGoogle Scholar |
Rice, CJ, Larson, ER, and Taylor, CA (2018). Environmental DNA detects a rare large river crayfish but with little relation to local abundance. Freshwater Biology 63, 443–455.
| Environmental DNA detects a rare large river crayfish but with little relation to local abundance.Crossref | GoogleScholarGoogle Scholar |
Roesma, DI, Djong, HT, Janra, MN, and Aidil, DR (2021). Freshwater vertebrates monitoring in Maninjau Lake, West Sumatra, Indonesia using environmental DNA. Biodiversitas Journal of Biological Diversity 22, 2794–2802.
| Freshwater vertebrates monitoring in Maninjau Lake, West Sumatra, Indonesia using environmental DNA.Crossref | GoogleScholarGoogle Scholar |
Rojahn, J, Gleeson, DM, Furlan, E, Haeusler, T, and Bylemans, J (2021). Improving the detection of rare native fish species in environmental DNA metabarcoding surveys. Aquatic Conservation: Marine and Freshwater Ecosystems 31, 990–997.
| Improving the detection of rare native fish species in environmental DNA metabarcoding surveys.Crossref | GoogleScholarGoogle Scholar |
Sato, H, Sogo, Y, Doi, H, and Yamanaka, H (2017). Usefulness and limitations of sample pooling for environmental DNA metabarcoding of freshwater fish communities. Scientific Reports 7, 14860.
| Usefulness and limitations of sample pooling for environmental DNA metabarcoding of freshwater fish communities.Crossref | GoogleScholarGoogle Scholar |
Schenekar, T, Schletterer, M, Lecaudey, LA, and Weiss, SJ (2020). Reference databases, primer choice, and assay sensitivity for environmental metabarcoding: lessons learnt from a re-evaluation of an eDNA fish assessment in the Volga headwaters. River Research and Applications 36, 1004–1013.
| Reference databases, primer choice, and assay sensitivity for environmental metabarcoding: lessons learnt from a re-evaluation of an eDNA fish assessment in the Volga headwaters.Crossref | GoogleScholarGoogle Scholar |
Seymour, M (2019). Rapid progression and future of environmental DNA research. Communications Biology 2, 80.
| Rapid progression and future of environmental DNA research.Crossref | GoogleScholarGoogle Scholar |
Sholihah, A, Delrieu-Trottin, E, Sukmono, T, Dahruddin, H, Risdawati, R, Elvyra, R, Wibowo, A, Kustiati, K, Busson, F, Sauri, S, Nurhaman, U, Dounias, E, Zein, MSA, Fitriana, Y, Utama, IV, Muchlisin, ZA, Agnèse, J-F, Hanner, R, Wowor, D, Steinke, D, Keith, P, Rüber, L, and Hubert, N (2020). Disentangling the taxonomy of the subfamily Rasborinae (Cypriniformes, Danionidae) in Sundaland using DNA barcodes. Scientific Reports 10, 2818.
| Disentangling the taxonomy of the subfamily Rasborinae (Cypriniformes, Danionidae) in Sundaland using DNA barcodes.Crossref | GoogleScholarGoogle Scholar |
Sholihah, A, Delrieu-Trottin, E, Condamine, FL, Wowor, D, Rüber, L, Pouyaud, L, Agnèse, J-F, and Hubert, N (2021a). Impact of pleistocene eustatic fluctuations on evolutionary dynamics in southeast asian biodiversity hotspots. Systematic Biology 70, 940–960.
| Impact of pleistocene eustatic fluctuations on evolutionary dynamics in southeast asian biodiversity hotspots.Crossref | GoogleScholarGoogle Scholar |
Sholihah, A, Delrieu-Trottin, E, Sukmono, T, Dahruddin, H, Pouzadoux, J, Tilak, M-K, Fitriana, Y, Agnèse, J-F, Condamine, FL, Wowor, D, Rüber, L, and Hubert, N (2021b). Limited dispersal and in situ diversification drive the evolutionary history of Rasborinae fishes in Sundaland. Journal of Biogeography 48, 2153–2173.
| Limited dispersal and in situ diversification drive the evolutionary history of Rasborinae fishes in Sundaland.Crossref | GoogleScholarGoogle Scholar |
Shu, L, Ludwig, A, and Peng, Z (2021). Environmental DNA metabarcoding primers for freshwater fish detection and quantification: in silico and in tanks. Ecology and Evolution 11, 8281–8294.
| Environmental DNA metabarcoding primers for freshwater fish detection and quantification: in silico and in tanks.Crossref | GoogleScholarGoogle Scholar |
Sigsgaard, EE, Carl, H, Møller, PR, and Thomsen, PF (2015). Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biological Conservation 183, 46–52.
| Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples.Crossref | GoogleScholarGoogle Scholar |
Sigsgaard, EE, Jensen, MR, Winkelmann, IE, Møller, PR, Hansen, MM, and Thomsen, PF (2020). Population-level inferences from environmental DNA – current status and future perspectives. Evolutionary Applications 13, 245–262.
| Population-level inferences from environmental DNA – current status and future perspectives.Crossref | GoogleScholarGoogle Scholar |
Simpfendorfer, CA, Kyne, PM, Noble, TH, Goldsbury, J, Basiita, RK, Lindsay, R, Shields, A, Perry, C, and Jerry, DR (2016). Environmental DNA detects critically endangered largetooth sawfish in the wild. Endangered Species Research 30, 109–116.
| Environmental DNA detects critically endangered largetooth sawfish in the wild.Crossref | GoogleScholarGoogle Scholar |
Stewart, KA (2019). Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodiversity and Conservation 28, 983–1001.
| Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA.Crossref | GoogleScholarGoogle Scholar |
Taberlet, P, Coissac, E, Pompanon, F, Brochmann, C, and Willerslev, E (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Molecular Ecology 21, 2045–2050.
| Towards next-generation biodiversity assessment using DNA metabarcoding.Crossref | GoogleScholarGoogle Scholar |
Thalinger, B, Wolf, E, Traugott, M, and Wanzenböck, J (2019). Monitoring spawning migrations of potamodromous fish species via eDNA. Scientific Reports 9, 15388.
| Monitoring spawning migrations of potamodromous fish species via eDNA.Crossref | GoogleScholarGoogle Scholar |
Trebitz, AS, Hoffman, JC, Darling, JA, Pilgrim, EM, Kelly, JR, Brown, EA, Chadderton, WL, Egan, SP, Grey, EK, Hashsham, SA, Klymus, KE, Mahon, AR, Ram, JL, Schultz, MT, Stepien, CA, and Schardt, JC (2017). Early detection monitoring for aquatic non-indigenous species: optimizing surveillance, incorporating advanced technologies, and identifying research needs. Journal of Environmental Management 202, 299–310.
| Early detection monitoring for aquatic non-indigenous species: optimizing surveillance, incorporating advanced technologies, and identifying research needs.Crossref | GoogleScholarGoogle Scholar |
Turner, CR, Uy, KL, and Everhart, RC (2015). Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biological Conservation 183, 93–102.
| Fish environmental DNA is more concentrated in aquatic sediments than surface water.Crossref | GoogleScholarGoogle Scholar |
Ulayya, N, Andayani, N, and Maryanto, AE (2020). Development of environmental DNA approaches to detect alligator gar (Atractosteus spatula) from water samples. IOP Conference Series: Earth and Environmental Science 481, 012013.
| Development of environmental DNA approaches to detect alligator gar (Atractosteus spatula) from water samples.Crossref | GoogleScholarGoogle Scholar |
Valiere, N, and Taberlet, P (2000). Urine collected in the field as a source of DNA for species and individual identification. Molecular Ecology 9, 2150–2152.
Wibowo, A, Wahlberg, N, and Vasemägi, A (2017). DNA barcoding of fish larvae reveals uncharacterised biodiversity in tropical peat swamps of New Guinea, Indonesia. Marine and Freshwater Research 68, 1079–1087.
| DNA barcoding of fish larvae reveals uncharacterised biodiversity in tropical peat swamps of New Guinea, Indonesia.Crossref | GoogleScholarGoogle Scholar |
Wibowo, A, Shibuno, T, and Sulit, VT (2018). The making of a center of excellence in science and technology on inland fisheries management: the SEAFDEC/IFRDMD. Fish for the People 16, 28–33.
Xiao, Y, Ouyang, Z, Xu, W, Xiao, Y, Zheng, H, and Xian, C (2016). Optimizing hotspot areas for ecological planning and management based on biodiversity and ecosystem services. Chinese Geographical Science 26, 256–269.
| Optimizing hotspot areas for ecological planning and management based on biodiversity and ecosystem services.Crossref | GoogleScholarGoogle Scholar |
Xiong, F, Shu, L, Zeng, H, Gan, X, He, S, and Peng, Z (2022). Methodology for fish biodiversity monitoring with environmental DNA metabarcoding: the primers, databases and bioinformatic pipelines. Water Biology and Security 1, 100007.
| Methodology for fish biodiversity monitoring with environmental DNA metabarcoding: the primers, databases and bioinformatic pipelines.Crossref | GoogleScholarGoogle Scholar |
Yachi, S, and Loreau, M (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proceedings of the National Academy of Sciences 96, 1463–1468.
| Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis.Crossref | GoogleScholarGoogle Scholar |
Yamanaka, H, and Minamoto, T (2016). The use of environmental DNA of fishes as an efficient method of determining habitat connectivity. Ecological Indicators 62, 147–153.
| The use of environmental DNA of fishes as an efficient method of determining habitat connectivity.Crossref | GoogleScholarGoogle Scholar |
Zhang, S, Zhao, J, and Yao, M (2020). A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish. Methods in Ecology and Evolution 11, 1609–1625.
| A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish.Crossref | GoogleScholarGoogle Scholar |


