Marine mammal strandings recorded in New Caledonia, South West Pacific Ocean, 1877 to 2022
Claire Garrigue A B * , Solène Derville
A B * , Solène Derville  A B C , Claire Bonneville A B , Maële Brisset B , Paco Bustamante
A B C , Claire Bonneville A B , Maële Brisset B , Paco Bustamante  D , Christophe Cleguer
D , Christophe Cleguer  E , Eric E. G. Clua F G , Willy Dabin H , Sylvie Fiat
E , Eric E. G. Clua F G , Willy Dabin H , Sylvie Fiat  A , Jean-Lou Justine
A , Jean-Lou Justine  I , Pauline Machful
I , Pauline Machful  J , Tepoerau Mai B K , Patrice Plichon L , Annie Portal J , Christine Sidobre A , Debbie Steel M , Jean-Christophe Vivier N and Elodie Vourey J
J , Tepoerau Mai B K , Patrice Plichon L , Annie Portal J , Christine Sidobre A , Debbie Steel M , Jean-Christophe Vivier N and Elodie Vourey J
A
B
C
D
E
F
G
H
I
J
K
L
M
N Deceased. Formerly of
Abstract
Strandings are an important source of information for estimating marine mammal biodiversity, particularly in data-sparse ocean basins such as Oceania.
Here, we report on knowledge acquired from 218 stranding events recorded in the waters of New Caledonia (1877–2022).
We investigated spatio-temporal distribution, stable isotope signatures, trace element concentrations, biometry measurements, genetic diversity, and diet, for the four most commonly stranded taxa (dugongs, 35% of events; sperm whales, 19%; Delphinidae, 18%; pygmy and dwarf sperm whales, 14%).
Beginning in 1991, reports of stranding events increased (183 events, 322 individuals, 20 species from seven families: Dugongidae, Physeteridae, Delphinidae, Kogiidae, Ziphiidae, Balaenopteridae, Otariidae), with hotspots identified on the west coast (Bourail, Ouano, Nouméa) and in Prony Bay. Causes of death were not determined in 84% of stranding events, but were identified in the majority of expert-led necropsies (24 of 29 individuals from 10 species). Yet, valuable information regarding the impact of anthropogenic activities was gathered for some species of concern, such as the endangered dugong (28% human-caused). Since 2016, training and outreach have been provided to rangers, veterinarians, and various public safety officers to support their engagement in the scientific monitoring of marine mammal strandings. A website (www.rescue.ird.nc) was developed to facilitate standardised data collection and storage, and to provide public access to stranding records.
Although the number of individuals reported here remains modest, this study provides new information on poorly documented species in New Caledonia.
Long-term monitoring of strandings can help design effective conservation measures.
Abstract
RESUME
Les échouages constituent une source d’informations importante pour estimer la biodiversité des mammifères marins, en particulier dans les bassins océaniques pour lesquels les données sont rares, comme l’Océanie
Nous présentons ici les connaissances acquises à partir de 218 échouages enregistrés dans les eaux de Nouvelle-Calédonie (1877–2022).
Nous avons étudié la distribution spatio-temporelle, les signatures isotopiques, les éléments traces, les mesures biométriques, la diversité génétique et le régime alimentaire des quatre taxons les plus fréquemment échoués (dugongs, 35% des échouages; cachalots, 19%; Delphinidae, 18% ; cachalots pygmées et nains, 14%).
À partir de 1991, les signalements d’échouages ont augmenté (183 événements, 322 individus, 20 espèces de sept familles: Dugongidae, Physeteridae, Delphinidae, Kogiidae, Ziphiidae, Balaenopteridae, Otariidae), avec des points chauds identifiés sur la côte ouest de la Grande Terre (Bourail, Ouano, Nouméa) et dans la Baie de Prony. Les causes de décès n’ont pas été déterminées pour 84% des échouages, mais elles ont été identifiées dans la majorité des autopsies réalisées par des vétérinaires (24 des 29 individus de 10 espèces). Des informations précieuses concernant l’impact des activités anthropiques ont été recueillies pour certaines espèces préoccupantes, telles que le dugong, une espèce en voie de disparition (28% des échouages d’origine humaine). Depuis 2016, des formations et des activités de sensibilisation ont été dispensées aux gardes nature, aux vétérinaires et à divers agents publics pour soutenir leur engagement dans la surveillance scientifique des échouages de mammifères marins. Un site Web (www.rescue.ird.nc) a été développé pour faciliter la collecte et le stockage de données standardisées et pour fournir un accès public aux enregistrements d’échouages.
Bien que le nombre d’individus signalés échoués ici reste modeste, cette étude apporte de nouvelles informations sur des espèces peu documentées en Nouvelle-Calédonie.
La surveillance à long terme des échouages peut aider à concevoir des mesures de conservation efficaces.
Keywords: age, diet, marine mammals, mtDNA, stable isotopes, stomach contents, stranding, trace elements.
Introduction
Monitoring marine mammal strandings is widely recognised as a valuable source of biological and ecological information on species otherwise difficult to observe at sea (Evans and Hammond 2004; Dalebout et al. 2005; Beasley et al. 2013). Close scientific monitoring of stranded marine mammals can provide low cost critical evidence of the threats faced by some populations, including disease outbreaks, incidental fisheries takes (‘bycatch’), shipstrikes, ingestion and entanglement in marine debris, and impacts of anthropogenic sounds (Laist et al. 2001; Cox et al. 2006; Meynecke and Meager 2016; Unger et al. 2016). The samples collected from stranded marine mammals can also be used to track pathogens, pollutants such as pesticides and industrial chemicals, particularly in the case of top predators, which can concentrate both organic and inorganic pollutants (Bustamante et al. 2003). Finally, stranding events can contribute to knowledge on local marine biodiversity (through species identification) and species genetic diversity and population structure. The knowledge gained from strandings is particularly important in the South Pacific, a vast area of tropical and subtropical ocean dotted with thousands of islands. Due to the productivity of their fringing coral reefs and the complex currents that these small landmasses generate, the waters surrounding islands may form an ‘oasis of life’ attracting marine mammal predators in these generally nutrient-poor seas (Menkes et al. 2015). Coral reefs encircling many South Pacific islands form shallow and sheltered lagoons that provide suitable habitat for marine mammals year-round (e.g. Indo-Pacific bottlenose dolphins, Tursiops aduncus (Bonneville et al. 2021)) and may also be used seasonally as breeding grounds for otherwise pelagic species (e.g. humpback whales, Megaptera novaeangliae (Garrigue et al. 2001)). Surveying such a vast area of ocean to study marine mammals, particularly cryptic, seasonal and/or more pelagic species can be time consuming and costly. Therefore, assessing strandings throughout the South Pacific’s islands allows us a glimpse of the species that inhabit these waters, and of their distribution.
New Caledonia is the largest archipelago in the South Pacific in terms of landmass, and is surrounded by a vast Economic Exclusive Zone (EEZ) spanning more than 1.3 million km2. Marine mammal strandings have been reported in this archipelago since as early as 1877, but have only been systematically monitored since the early 1990s (with a first summary of strandings between 1877 and 2005 provided by Borsa 2006). These stranding events have considerably contributed to the knowledge of local dugong (Dugong dugon) and cetacean populations. For instance, to date eight cetacean species, or subspecies, were first recorded in New Caledonia from stranding events – a pygmy and a dwarf sperm whale in single strandings in 1972 and 1974 (Kogia breviceps and K. sima; (Robineau and Rancurel 1981; Sylvestre 1988), a pygmy blue whale (Balaenoptera musculus brevicauda) single stranding in 2002 (Clua 2002; Garrigue et al. 2003; Borsa and Hoarau 2004), single stranding of common dolphin (Delphinus delphis) around 1877, sei whale (Balaenoptera borealis) in 1962, Antarctic minke whale (Balaenoptera bonaerensis) in 1993, melon-headed whale (Peponocephala electra) in 2003 and New Zealand fur seal (Arctocephalus forsteri) in 1972 (all in Borsa (2006)). In addition, two cetacean species were first reported in New Caledonia from mass-stranding events: pygmy killer whales (Feresa attenuata, (Clua et al. 2014)) and Longman’s beaked whales (Indopacetus pacificus, (Garrigue et al. 2016)). These events have led to improved knowledge of these species in New Caledonia and the South Pacific at large.
In this study, we present a detailed summary of all marine mammal stranding events reported in New Caledonia since 1877. In addition to describing species identification, the general trends in spatio-temporal distribution, and causes of death, we present insights gained from analysing mitochondrial DNA (mtDNA), stomach contents, age, stable isotope values, and trace element concentrations from stranded individuals.
Methods
Study area
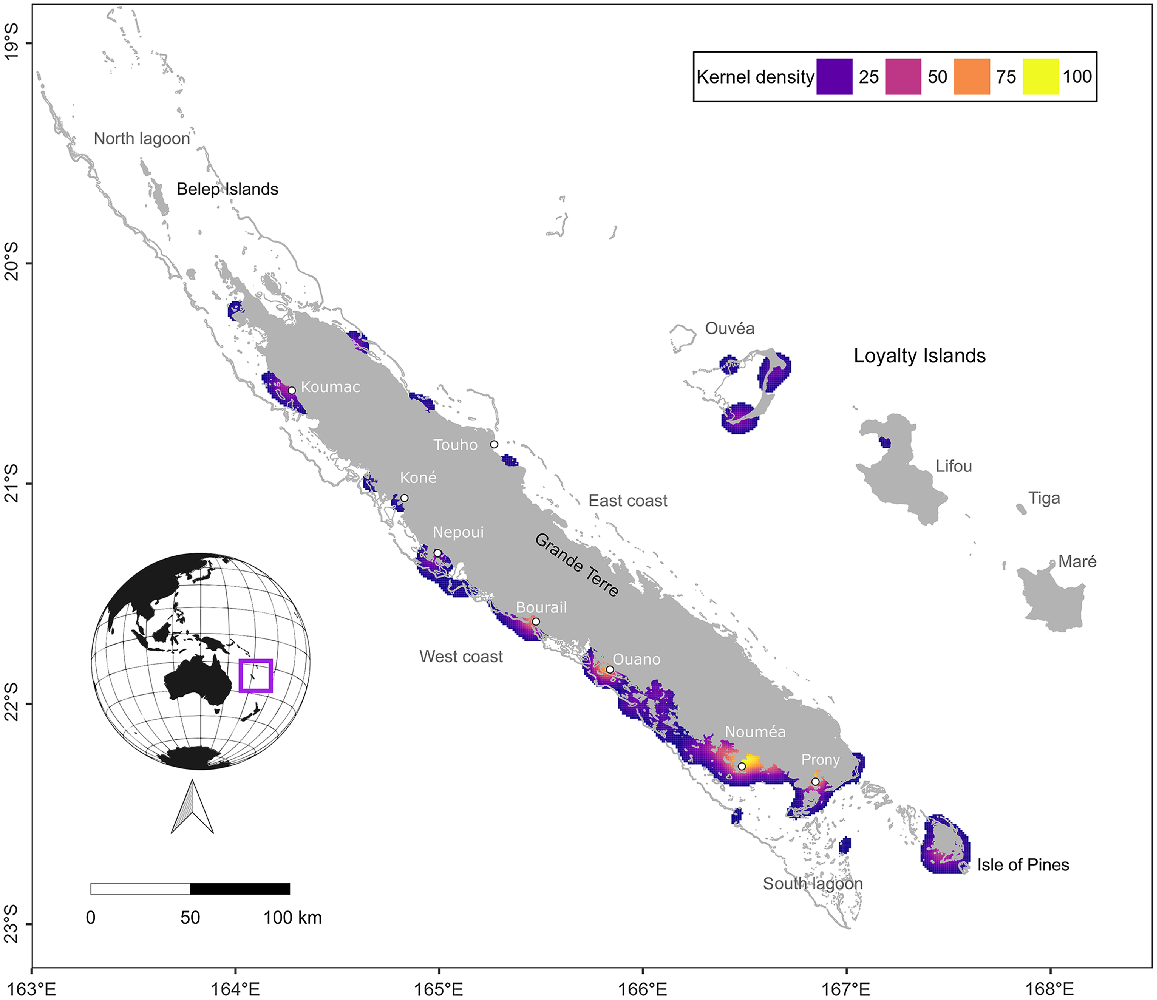
The main island of New Caledonia, Grande Terre (a fragment of ancient Gondwana), is 400 km long by 50 km wide, and is surrounded by an immense barrier reef 1600 km in length, which forms large lagoons with waters up to 70 m deep and numerous small islets (Fig. 1). Three islands are included in these lagoons: Isle of Pine in the south and the Belep Islands (Art and Pott) in the north. Outside of the barrier reef, the waters drop quickly from a few hundred to more than 3000 m. The Loyalty Islands (Ouvea, Tiga, Lifou, and Maré) lie approximately 100 km to the east of Grande Terre, whereas the uninhabited Chesterfield–Bellona coral reef complex is located approximately 600 km to the west of Grande Terre. The study area extends over the entire New Caledonian archipelago with the exception of the Chesterfield–Bellona coral reef complex (not shown in maps) where no strandings were reported (Fig. 1).
Kernel density estimates of all recorded marine mammal strandings in New Caledonia between 1991 and 2022. Density values below 10% are not shown. Orange colours indicate higher densities and blue colours indicate lower densities. Land and shallow reefs are shown in grey. Note that stranding records are likely to be subject to a spatial bias as 76% of the 238 000 inhabitants of New Caledonia are found in Nouméa and its surroundings.
Data collection
Strandings are defined as dead or live animals that were either found washed ashore, floating nearshore, or by-caught in fishing gear. In most cases, rescue was attempted when animals stranded alive. For this study, the term mass stranding refers to two or more animals stranded together, with the exception of mother–calf pairs that are considered as isolated stranding events. The number of stranding events and stranded individuals reported in this paper were summarised by family and species. The first necropsy was conducted by a veterinarian in 1994 and continues to be conducted on a portion of stranded animals until the present day.
Information on stranding events before 1991 was sourced mainly from local newspapers, discussion with local people and published papers (Rancurel 1973; Robineau and Rancurel 1981; Sylvestre 1988; Borsa 2006, 2022). Since 1991, the marine mammal specialist, local nongovernmental organisation Operation Cétacés has attempted to collect information on all stranding events occurring in New Caledonian waters. Since 2016, the lead author has recommended standardised protocols for the collection of data and biological samples and initiated the training of persons authorised to intervene in stranding events, such as rangers from the environmental services of the three provinces of New Caledonia and veterinarians. Due to remote locations, some information such as exact date, geographical location or biological data, was obtained weeks or months later resulting in variable levels of uncertainty in some stranding records.
When examination of the carcass was possible, data were collected following guidelines set up by the French national stranding network, depending on the degree of decay (https://www.observatoire-pelagis.cnrs.fr/echouages/reseau-national-echouage/). Given that complete data collection was not always possible, we recommended at least noting the date, the precise location of the stranding (with geographical coordinates if possible), the state of decomposition of the carcass, the presence of scars that may indicate human interaction, the total length of the animal in order to estimate its age-class (Jefferson et al. 2008; Lanyon et al. 2021), photographs, and collection of a skin sample. When possible, teeth, stomach contents, parasites, and tissues (e.g. skin, muscle, liver, kidney) were collected and complete morphological measurements were made. If a veterinarian was available, a necropsy could be performed.
Spatial and temporal analysis
All marine mammal stranding locations available were mapped and analysed using R statistical software (R Core Team 2023). The total number of strandings was analysed annually and monthly. Among the species identified with good confidence based on visual field identification and/or genetic identification (certain and probable), we also made separate analyses for the most commonly stranded taxa: dugongs, short-finned pilot whales (Globicephala macrorhynchus), dwarf and/or pygmy sperm whales, and sperm whales (Physeter macrocephalus). Spatial distribution was assessed with kernel density estimates applied to stranding events that occurred between 1991 and 2022 using the R package MASS (Ver. 7.3). Based on the dimensions of the archipelago and the lagoon width, kernels were applied with a bandwidth of 20 km and at a resolution of 2 km. Kernel estimates and observations were represented over maps of land and coral reefs provided by Andréfouët et al. (2008). The number of individuals per stranding was used as weights. Temporal distribution across years was investigated with generalised linear models (GLM) in which the number of stranding events was modelled with a Poisson distribution as a function of year. Temporal distribution of stranding events throughout the year was investigated with χ2 tests applied to the number of events per austral season: spring (October–December), summer (January–March), fall (April–June), and winter (July–September).
Biological analysis
Genetic analyses were used to (1) confirm species identity (and sex) especially if the carcass was very decomposed, or if it was a rare species, and, (2) for a subset of species, determine the population of origin. For the first objective, mtDNA control region sequences (see Supplementary Material S1 for details on lab procedure) were checked against Genbank (https://www.ncbi.nlm.nih.gov/genbank/) using a nucleotide Basic Local Alignment Search Tool (BLAST) search. For the second objective, mtDNA control region sequences generated as part of this study were aligned to previously published sequences downloaded from GenBank (Table S1). GenBank sequences were chosen to be part of a reference dataset if they contained no uncertainties and if the sampling location was known in sequence details or in the related publication. The geographic coverage and number of sequences contained in these reference datasets varied depending on the species of interest. Details of each dataset used are available in Table S2. For sperm whales, three sequences originated from biopsy samples taken on live animals were also included in the analyses. All sequences are available on GenBank: https://www.ncbi.nlm.nih.gov/genbank/ (Genbank accession #OQ736578-OQ736596).
Given that haplotype frequencies were unknown for many of the sequences that comprise the reference datasets, only one sequence per haplotype per region was used for this study. Consequently, only likely population origin was explored and no population differentiation analyses were conducted. For each species, a median-joining haplotype network of relationships among the worldwide mtDNA haplotypes was created using Network V10 (Fluxus Technology).
For cetacean and dugong teeth, Growth Layer Groups (GLGs) were counted, assuming that one GLG equals 1 year (Gurevich et al. 1980; Perrin and Myrick 1980; Klevezal 1996; Lockyer and Garrigue 2021), see Supplementary Material 1 for details on the procedure). For cetaceans, at least two different readers made three independent readings of three tooth sections for each individual, thus resulting in over 18 independent age determinations per individual. All readings were recorded to the nearest whole year and age was expressed as mean ± s.d. for each individual. Inter-reader variation in age determination was assessed by using a paired t-test, whereas precision in age reading was compared across the age series by applying a χ2 test comparing errors in age determination among animals of 0–9, 10–15 and 16–23 years. For dugong, the upper posterior incisors, called more commonly the ‘tusks’, were used for ageing as they are their only permanent teeth. The tusks predominantly erupt in males, occasionally in females, and hence result in only a minimum age in quite old animals (Mitchell 1978; Marsh 1980, 2009). The GLGs were counted using the naked eye and a magnifying glass. Two people examined each section independently three times.
Prey items were sorted into categories (fish, cephalopods, crustaceans and gelatinous plankton), identified to the lowest possible taxonomic level based on literature, and classified according to degree of digestion (fresh, slightly digested, advanced digestion, hard parts) and developmental stage (larvae, juvenile, adult, see Supplementary Material 1 for details on lab procedure). The data were analysed to highlight proportions of prey items without hard parts (cephalopod’s beaks) by number (%N, percentage representing the total prey items from a particular taxon identified), proportions by weight (%W, percentage representing the total weight of remains from a particular taxon), and frequencies of occurrence (%FO, percentage of stomachs containing a particular taxon). The vertical distributions of the taxa identified in the stomachs were characterised according to the literature and from data obtained during research collaborations between the Fisheries Aquaculture and Marine Ecosystem Laboratory (FAME) of the Pacific Community (SPC, Noumea) and the French National Research Institute for Sustainable Development (IRD), conducted in the EEZ of New Caledonia (Allain and Menkes 2011, 2021; Garrigue and Derville 2019; Olu and Allain 2020).
Samples used for trace element, radionuclide and stable isotope analyses were freeze-dried and ground with a porcelain mortar and pestle (Garrigue et al. 2000; Bustamante et al. 2001, 2003). Powdered dried tissues were stored in polyethylene vials prior to analysis (see Supplementary Material 1 for details on lab procedure).
Fourteen trace elements were analysed: silver (Ag), arsenic (As), cadmium (Cd), cobalt (Co), chromium (Cr), copper (Cu), iron (Fe), mercury (Hg), manganese (Mn), nickel (Ni), lead (Pb), selenium (Se), vanadium (V) and zinc (Zn) as described elsewhere (Kojadinovic et al. 2011). Trace element concentrations are expressed in μg.g−1 of dry weight (dw).
Stable isotope analyses were conducted to obtain information on the feeding habitats of the animals (δ13C), and their trophic position in the food webs (δ15N). As the turnover time of these elements varies by tissue type, from several days (e.g. liver) to several months (e.g. muscle), stable isotopes were analysed between the available tissues to cover a wide temporal range. Results are expressed in parts per thousand (‰) in the usual δ notation, relative to Vienna Pee Dee Belemnite for δ13C and atmospheric N2 for δ15N, following the formula:
where R is 13C/12C or 15N/14N, respectively. The measurement error was <0.15 ‰ for both δ13C and δ15N values.
Results and discussion
General trends
A total of 218 marine mammal stranding events, accounting for a minimum of 409 individuals, were reported around New Caledonia between 1877 and 2022 (Table S3, Fig. S1). As a result of both field-based, photographic, and genetic identification, species were identified with certainty in 82% of the cases (n = 179), and probable in 13% (n = 28) of the cases. The species could not be identified in 5% (n = 11) of the stranding events, and thus were considered as ‘unknown’. Of the 30 species known to have used New Caledonian waters, 22 were documented in stranding events (one sirenian, one pinniped, and 20 cetaceans, including five baleen whales and 15 toothed whales (Garrigue 2007; Garrigue and Poupon 2013)). Of these 22, six have never been documented alive in the waters of New Caledonia and are only known to use the region from these stranding events (pygmy blue whale, pygmy killer whale, striped dolphin (Stenella coeruleoalba), common dolphin, melon-headed whale, rough-toothed dolphin (Steno bredanensis). Most of the stranding events (84%, n = 183 events involving a minimum of 322 individuals of 20 species) occurred during the period 1991–2022. Between 1996 and 2020, 29 individuals (from 10 species) were necropsied with the support of a veterinarian (Table S4).
The majority of events between 1877 and 2022 (n = 192 or 88%) were considered single strandings, including three mother–calf pairs. Twenty six events (12%) were considered mass strandings with a minimum of two and a maximum of 50 individuals. Short-finned pilot whales were the most common species to mass strand, comprising 42% (n = 11) of all events, and 31% (n = 122) of the identified individuals, with the remaining 58% of events involving eight different species (striped dolphin, spinner dolphin (Stenella longirostris), false killer whale (Pseudorca crassidens), pygmy killer whale, Longman’s beaked whale, pygmy and dwarf sperm whales, sperm whale). The most common taxa to strand were dugongs, comprising 31% (n = 65) of the documented stranding events for the whole study period with the majority of those 35% (n = 60) between 1991 and 2022, followed by sperm whales (22%, n = 45; 1991–2022: 19%, n = 33), Delphinidae (18%, n = 37; 1991–2022: 18%, n = 31) and Kogiidae (14%, n = 28; 1991–2022: 14%, n = 25). Delphinidae had the highest number of individuals stranded, (52%, n = 208, 1991–2022: 50%, n = 152) due to the mass strandings of pilot whales, which account for 59% (n = 122; 1991–2022: 47%, n = 72) of the stranded Delphinidae individuals.
State of carcasses (1991–2022)
No information on the state of carcasses was available for 32 of the 322 individuals that stranded between 1991 and 2022. In 35 events, all or some of the animals (n = 130, 40%) stranded alive. Of these, 92 individuals were refloated. The species that stranded alive were dugong, pygmy and dwarf sperm whales, false killer whale, pygmy killer whale, short-finned pilot whale, New Zealand fur seal, spinner dolphin, Longman’s beaked whale, and Antarctic minke whale. For the remaining 185 carcasses, 40% were freshly dead, 43% more or less decomposed, and 17% were highly decomposed, with only fragments remaining (Table S3).
Causes of stranding (1991–2022)
The cause of stranding was unknown for most of the individuals (n = 271, 84%) because (1) no trained observers could access the carcass, (2) the carcass was too deteriorated to be examined, or the individual was refloated (34%, n = 92). When identified (n = 51), the cause of death for 55% of individuals was from natural causes (e.g. predation, disease, starvation, meteorological events such as following hurricanes, see Clua et al. 2014), and was of anthropogenic origin for 45% of the individuals (e.g. intentionally killed 20% (n = 10), boat collision 18% (n = 9), entanglement in fishing gear 8% (n = 4)). In rare cases, marine debris, such as plastics, were found in stomachs but did not necessarily cause death (see Table S4). In three cases of necropsies followed by laboratory analysis, pathogens such as morbillivirus and poxvirus were identified as putative killing agents for Longman’s beaked whale, striped and Indo-Pacific bottlenose dolphins (Table S4).
Among the deaths of anthropogenic origin, all but one of the intentional takes were observed in dugongs (see below, dugong focus taxa). The one other case was an Indo-Pacific bottlenose dolphin showing various wounds caused by weapons, recorded in 1994. Moreover, within the stranding records six dugongs (Tables S3 and S4) and one sperm whale showed clear evidence of propeller injuries or trauma linked to boat collisions. In addition, alleged collisions between a humpback whale and high-speed vessels were reported in the local newspaper in February 1998 and August 2004 but no biological evidence could be collected. Finally, a photographic analysis of Indo-Pacific bottlenose dolphin dorsal fin injuries revealed that many individuals in the south lagoon have been subject to propeller hits (Bonneville et al. 2021). Both lagoon and pelagic species were entangled in fishing gear with two dugongs entangled in fishing nets, one sperm whale entangled in a fixed fish-aggregating device used by professional coastal fishermen or recreational fishermen, and one young unweaned short-finned pilot whale caught in a long line fishing rope.
Temporal and spatial distribution
Reported stranding events increased in New Caledonia over the entire modern period from 1962 onwards (Fig. 2; GLM year effect: z = 9.88, P < 0.001, deviance explained = 0.60) and during the more recent study period 1991–2022 (z = 3.85, P < 0.001, deviance explained = 0.35). Events per year ranged from two in 1991 to a maximum of 15 in 2020 (mean ± s.d. 6 ± 3 events/year). Stranding occurred year-round, with no significant effect of season detected in events since 1877 (χ2 test: χ2 = 5.4, P = 0.139) nor since 1991 (χ2 test: χ2 = 4.2, P = 0.251).
Temporal distribution of stranding events in New Caledonia between 1877 and 2022. Historical period represented on the histogram ranges up to 1961 (dotted line). The recent study period with consistent stranding monitoring starts in 1991 (dashed line).
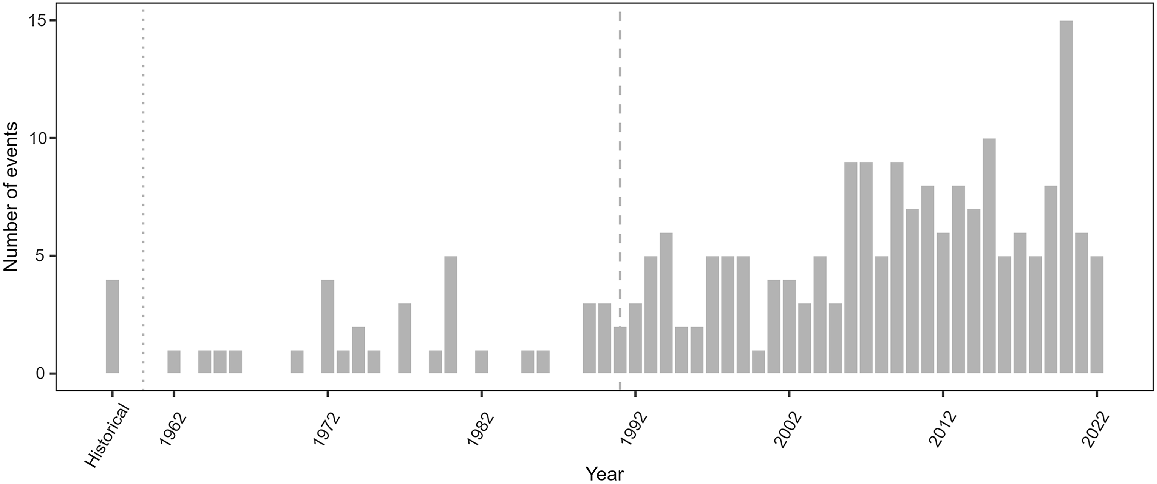
The increase in annual number of stranding events since 1962 is largely due to an increase in detection as a consequence of greater survey effort and public awareness. Stranding records from the end of the 19th and during most of the 20th century were retrieved from opportunistic sightings and newspapers and it is highly probable that most events were undetected or unreported. Such sampling bias could also have contributed to the observed stranding increase since 1991, although over this period monitoring was much more consistent and a real increase in strandings may have occurred.
Stranding events were reported all around the mainland as well as all four of the Loyalty Islands, Isle of Pines, and Belep islands, along the coastlines as well as on the reefs surrounding the archipelago. The majority of strandings were located on the west coast of the mainland (Fig. 1), consistent with aerial survey observations of a greater presence of marine mammals on the west coast compared to the east coast of the mainland (Garrigue et al. 2008; Van Canneyt et al. 2016; Cleguer et al. 2017; Laran et al. 2023). Stranding hotspots were identified around Bourail, Ouano, Nouméa, and Prony Bay. However, as with the temporal trends described above, the spatial distribution of stranding records is also likely affected by sampling bias and we do not know how far carcasses floating in the lagoon can drift from the moment of death to stranding of the body on the shore (Peltier et al. 2012). Three quarters of the New Caledonian human population is concentrated in or near Noumea (182 000 inhabitants in 2019, equivalent to 76% of the total New Caledonian archipelago population, INSEE data https://www.insee.fr/fr/statistiques/2122859), which may explain why more strandings were reported in this area. In comparison, all other counties or towns represented in Fig. 1 have similarly small population sizes (e.g. Bourail ~5500 people in 2019, Koumac ~ 4000 people, Thio ~2500 people). Despite not having high permanent human population densities, other stranding hotspots such as Ouano, Bourail, and Prony Bay are relatively popular tourist destinations and are known to host greater numbers of some marine mammal species (humpback whales in Prony Bay (Derville et al. 2019), dugongs in Bourail and Ouano (Cleguer et al. 2015)). Prony Bay also appears to be a hotspot of stranding diversity of both coastal and pelagic marine mammal species (five different species recorded stranded), which may be due to the bay being largely open to the ocean.
Stable isotopes and trace elements
There was wide variation of stable isotope values, with a clear segregation between dugong (ranging from −6.3 to −9.8 ‰ for δ13C and from 2.4 to 5.5 ‰ for δ15N) and toothed whales (ranging from −15.6 to −18.8 ‰ for δ13C and from 11.0 to 15.0 ‰ for δ15N) independent of the tissues analysed (Table 1). Stable isotopes were strongly correlated between liver and muscle (r2 = 0.961 for δ13C and r2 = 0.935 for δ15N) and between liver and kidney (r2 = 0.950 in both cases).
| Taxa | Code of individual | Tissue | δ13C | δ15N | Ag | As | Cd | Co | Cr | Cu | Fe | Hg | Mn | Ni | Pb | Se | V | Zn | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F. attenuata | EC2006-02-02 | Liver | −16.3 | 13.9 | 7.10 | <dl | 11.9 | 0.08 | <dl | 15 | 902 | 1288 | 13 | <dl | 0.29 | 466 | <dl | 83 | |
| F. attenuata | EC2006-04-01 | Liver | −17.1 | 13.4 | 10.2 | <dl | 22.5 | 0.06 | <dl | 26 | 1045 | 1075 | 9.7 | <dl | 0.27 | 412 | <dl | 98 | |
| F. attenuata | EC2006-04-02 | Liver | −17.0 | 14.5 | 6.28 | <dl | 28.7 | 0.05 | <dl | 32 | 1135 | 1645 | 12 | <dl | 0.47 | 551 | <dl | 93 | |
| F. attenuata | EC2006-04-03 | Liver | −16.4 | 12.8 | 2.72 | <dl | 6.42 | 0.05 | <dl | 18 | 575 | 147 | 9.4 | <dl | 0.18 | 59.5 | <dl | 90 | |
| F. attenuata | EC2006-07-01 | Liver | −17.6 | 14.8 | 12.1 | <dl | 19.8 | 0.09 | <dl | 24 | 1592 | 3959 | 8.8 | <dl | 0.19 | 1314 | <dl | 115 | |
| Mean | −16.9 | 13.9 | 7.68 | 17.9 | 0.07 | 23 | 1050 | 1623 | 11 | 0.28 | 561 | 96 | |||||||
| s.d. | 0.5 | 0.8 | 3.63 | 8.8 | 0.02 | 7 | 370 | 1418 | 2 | 0.12 | 461 | 12 | |||||||
| G. macrorhynchus | EC1997-01-01 A | Liver | −18.8 | 13.1 | ND | ND | 225 | <dl | <dl | 37 | 1472 | 1411 | 7.1 | <dl | ND | 627 | 0.06 | 136 | |
| G. macrorhynchus | EC1997-01-02 A | Liver | −18.3 | 14.4 | ND | ND | 464 | 0.04 | <dl | 51 | 1535 | 1452 | 6.8 | <dl | ND | 758 | 0.10 | 113 | |
| G. macrorhynchus | EC2009-07-01 | Liver | −17.2 | 12.1 | 3.59 | 2.8 | 226 | 0.08 | 0.68 | 30 | 1340 | 334 | 6.9 | 0.13 | 0.10 | 192 | <dl | 185 | |
| G. macrorhynchus | EC2009-07-02 | Liver | −17.2 | 12.0 | 3.96 | 3.2 | 363 | 0.07 | 0.22 | 29 | 1035 | 587 | 8.7 | 0.08 | 0.19 | 422 | <dl | 142 | |
| G. macrorhynchus | EC2009-07-03 | Liver | −16.8 | 11.7 | 3.26 | 1.9 | 198 | 0.06 | 0.24 | 23 | 2290 | 188 | 5.2 | <dl | 0.22 | 188 | <dl | 114 | |
| Mean | −17.6 | 12.7 | 3.60 | 2.6 | 295 | 0.07 | 0.38 | 34 | 1535 | 794 | 6.9 | 0.11 | 0.17 | 437 | 0.08 | 138 | |||
| s.d. | 0.8 | 1.1 | 0.35 | 0.7 | 114 | 0.02 | 0.26 | 11 | 464 | 599 | 1.3 | 0.04 | 0.07 | 255 | 0.03 | 29 | |||
| I. pacificus | EC2013-06-01 B | Liver | −17.0 | 13.1 | 1.57 | 18 | 97 | 0.34 | 0.13 | 10 | 1525 | 131 | 5.3 | <dl | <dl | 57.7 | <dl | 161 | |
| I. pacificus | EC2013-06-02 B | Liver | −16.7 | 13.8 | 2.25 | 24 | 142 | 0.37 | 0.20 | 13 | 1422 | 180 | 6.0 | <dl | <dl | 103 | <dl | 168 | |
| I. pacificus | EC2013-06-03 B | Liver | −16.9 | 12.6 | 2.51 | 23 | 169 | 0.24 | 0.15 | 10 | 924 | 800 | 8.3 | <dl | <dl | 278 | <dl | 181 | |
| Mean | −16.9 | 13.1 | 2.11 | 22 | 136 | 0.32 | 0.16 | 11 | 1290 | 370 | 6.5 | 146 | 170 | ||||||
| s.d. | 0.1 | 0.6 | 0.49 | 3.3 | 36 | 0.07 | 0.04 | 2 | 321 | 373 | 1.6 | 117 | 10 | ||||||
| K. breviceps | EC1997-04-01 A | Liver | −15.9 | 14.4 | ND | ND | 28.8 | <dl | <dl | 8 | 2503 | 7.86 | 5.2 | <dl | ND | 20.7 | 0.10 | 52 | |
| K. breviceps | EC1997-03-01 A | Liver | −17.3 | 14.5 | ND | ND | 47.5 | 0.05 | <dl | 18 | 3120 | 77.3 | 5.0 | <dl | ND | 25.2 | 0.68 | 54 | |
| K. breviceps | EC2007-05-01 | Liver | −16.8 | 12.5 | 0.46 | <dl | 0.04 | 0.15 | 1.53 | 25 | 1649 | 1.15 | 2.7 | <dl | <dl | 14.7 | <dl | 40 | |
| K. breviceps | EC2007-04-01 | Liver | −17.4 | 13.0 | 1.31 | <dl | 17.1 | 0.27 | <dl | 13 | 1140 | 50.5 | 4.8 | <dl | <dl | 30.7 | <dl | 79 | |
| K. breviceps | EC2007-03-01 | Liver | −17.0 | 13.5 | 1.12 | <dl | 14.7 | 0.11 | <dl | 9 | 2389 | 33.8 | 2.5 | <dl | <dl | 25.0 | <dl | 51 | |
| K. breviceps | EC2006-09-01 | Liver | −17.0 | 12.8 | 0.58 | <dl | 12.0 | 0.25 | 1.80 | 6 | 2434 | 27.7 | 2.2 | 1.78 | <dl | 29.9 | <dl | 43 | |
| K. breviceps | EC2010-04-01 | Liver | −17.1 | 12.6 | 0.33 | <dl | 7.6 | 0.18 | 0.38 | 5 | 2138 | 20.7 | 2.8 | 1.26 | <dl | 25.8 | 0.36 | 44 | |
| Mean | −16.9 | 13.3 | 0.76 | 18.2 | 0.17 | 1.24 | 12 | 2196 | 31.3 | 3.6 | 1.52 | 24.6 | 0.38 | 52 | |||||
| s.d. | 0.5 | 0.9 | 0.43 | 15.6 | 0.08 | 0.75 | 7 | 640 | 26.1 | 1.3 | 0.37 | 5.5 | 0.29 | 13 | |||||
| K. sima | EC2011-01-01 | Liver | −17.3 | 11.5 | 0.47 | 1.4 | 21.1 | 0.13 | 1.06 | 8 | 2556 | 25.3 | 6.6 | 0.66 | 0.02 | 23.2 | <dl | 86 | |
| D. dugon | EC2006-01-01 | Liver | −7.3 | 5.4 | 0.46 | <dl | 0.69 | 45.9 | <dl | 7 | 338 | 2.65 | 4.8 | 0.49 | <dl | 17.1 | <dl | 774 | |
| D. dugon | EC2006-13-01 | Liver | −7.3 | 5.5 | 0.43 | 16 | 2.34 | 36.7 | 1.45 | 9 | 1171 | 1.27 | 1.5 | <dl | <dl | 16.7 | <dl | 2389 | |
| D. dugon | EC2009-01-01 | Liver | −9.8 | 2.8 | 0.29 | 7.2 | 1.06 | 38.1 | 1.29 | 8 | 8958 | 0.16 | 1.9 | 0.71 | 0.09 | 1.63 | <dl | 2880 | |
| D. dugon | EC2011-05-01 | Liver | −6.4 | 2.9 | 1.81 | 0.3 | <dl | 4.63 | 0.45 | 239 | 1548 | 0.05 | 2.9 | 0.79 | 0.03 | 2.28 | <dl | 666 | |
| Mean | −7.7 | 4.1 | 0.75 | 7.7 | 1.36 | 31.3 | 1.06 | 66 | 3004 | 1.03 | 2.8 | 0.66 | 0.06 | 9.42 | 1677 | ||||
| s.d. | 1.5 | 1.5 | 0.71 | 7.7 | 0.86 | 18.2 | 0.53 | 115 | 4002 | 1.21 | 1.5 | 0.16 | 0.04 | 8.63 | 1124 | ||||
| F. attenuata | EC2006-02-02 | Kidney | −16.1 | 13.2 | 0.58 | <dl | 25.3 | 0.05 | <dl | 14 | 624 | 179 | 3.1 | <dl | 0.10 | 83.0 | <dl | 69 | |
| F. attenuata | EC2006-04-01 | Kidney | −16.1 | 13.5 | 0.94 | <dl | 27.7 | 0.06 | <dl | 11 | 472 | 249 | 1.8 | <dl | 0.10 | 96.1 | <dl | 69 | |
| F. attenuata | EC2006-04-02 | Kidney | −16.2 | 14.6 | 0.94 | <dl | 68.4 | 0.05 | <dl | 12 | 1031 | 293 | 2.9 | <dl | 0.17 | 124 | <dl | 77 | |
| F. attenuata | EC2006-04-03 | Kidney | −16.1 | 14.8 | 0.45 | <dl | 30.6 | 0.07 | <dl | 16 | 385 | 128 | 2.9 | <dl | 0.53 | 69.1 | <dl | 97 | |
| F. attenuata | EC2006-07-01 | Kidney | −16.7 | 15.0 | 1.94 | <dl | 46.2 | 0.45 | <dl | 18 | 1718 | 650 | 4.7 | <dl | 0.12 | 247 | <dl | 108 | |
| Mean | −16.2 | 14.2 | 0.97 | 39.7 | 0.14 | 14 | 846 | 300 | 3.1 | 0.20 | 124 | 84 | |||||||
| s.d. | 0.3 | 0.8 | 0.58 | 18.0 | 0.17 | 3 | 547 | 206 | 1.0 | 0.19 | 72 | 17 | |||||||
| G. macrorhynchus | EC2009-07-01 | Kidney | −16.3 | 13.1 | 0.40 | 3.1 | 343 | 0.16 | 0.56 | 15 | 612 | 90 | 3.4 | 0.12 | 0.03 | 63.0 | <dl | 142 | |
| G. macrorhynchus | EC2009-07-02 | Kidney | −16.2 | 13.0 | 0.29 | 2.7 | 332 | 0.07 | 0.68 | 9 | 458 | 61 | 2.9 | 0.20 | 0.03 | 45.1 | <dl | 104 | |
| G. macrorhynchus | EC2009-07-03 | Kidney | −16.2 | 12.8 | 0.16 | 2.9 | 278 | 0.08 | 2.07 | 10 | 476 | 32 | 2.8 | 0.15 | 0.02 | 30.3 | <dl | 102 | |
| Mean | −16.2 | 13.0 | 0.28 | 2.9 | 318 | 0.10 | 1.10 | 11 | 515 | 61 | 3.0 | 0.16 | 0.03 | 46.2 | 116 | ||||
| s.d. | 0.1 | 0.2 | 0.12 | 0.2 | 35 | 0.05 | 0.84 | 3 | 84 | 29 | 0.3 | 0.04 | 0.01 | 16.4 | 22 | ||||
| I. pacificus | EC2013-06-01 B | Kidney | −17.4 | 13.3 | <dl | 2.3 | 216 | 0.27 | 1.34 | 8 | 941 | 128 | 1.8 | 1.59 | <dl | 15.3 | <dl | 102 | |
| I. pacificus | EC2013-06-02 B | Kidney | −16.4 | 13.7 | 1.89 | 3.0 | 223 | <dl | 0.20 | 7 | 637 | 12.0 | 1.1 | <dl | <dl | 10.0 | <dl | 81 | |
| I. pacificus | EC2013-06-03 B | Kidney | −17.2 | 12.7 | 6.92 | 19 | 146 | 0.23 | 0.13 | 11 | 764 | 620 | 8.3 | 0.43 | <dl | 346 | <dl | 169 | |
| Mean | −17.0 | 13.2 | 4.41 | 8.2 | 195 | 0.25 | 0.56 | 9 | 781 | 253 | 3.7 | 1.01 | 124 | 117 | |||||
| s.d. | 0.6 | 0.5 | 3.56 | 9.6 | 42.9 | 0.03 | 0.68 | 2 | 153 | 323 | 4.0 | 0.82 | 193 | 46 | |||||
| K. breviceps | EC2007-05-01 | Kidney | ND | ND | <dl | <dl | 0.04 | 0.08 | 1.67 | 8 | 1149 | 0.73 | 2.0 | <dl | <dl | 17.6 | <dl | 63 | |
| K. breviceps | EC2007-04-01 | Kidney | ND | ND | <dl | <dl | 136 | 0.11 | <dl | 7 | 511 | 14.7 | 2.3 | <dl | <dl | 24.5 | <dl | 75 | |
| K. breviceps | EC2007-03-01 | Kidney | ND | ND | 0.26 | <dl | 47.2 | 0.13 | <dl | 6 | 906 | 62.2 | 1.4 | <dl | <dl | 43.0 | <dl | 54 | |
| K. breviceps | EC2006-09-01 | Kidney | −17.0 | 13.6 | 0.11 | <dl | 56.4 | 0.11 | <dl | 6 | 815 | 37.9 | 1.6 | <dl | <dl | 55.8 | <dl | 84 | |
| K. breviceps | EC2010-04-01 | Kidney | −16.9 | 13.0 | 0.08 | <dl | 97.4 | 0.11 | 0.52 | 7 | 1036 | 17.4 | 2.2 | <dl | 0.04 | 43.6 | <dl | 73 | |
| Mean | −16.9 | 13.3 | 0.15 | 67.4 | 0.11 | 7 | 884 | 26.6 | 1.9 | 0.04 | 36.9 | 70 | |||||||
| s.d. | 0.0 | 0.4 | 0.09 | 51.6 | 0.02 | 1 | 244 | 23.9 | 0.4 | 15.5 | 12 | ||||||||
| K. sima | EC2011-01-01 | Kidney | −16.9 | 11.8 | 0.10 | 2.9 | 30.8 | 0.18 | 2.21 | 10 | 573 | 36.1 | 3.2 | 0.65 | 0.04 | 54.3 | <dl | 92 | |
| D. dugon | EC2009-01-01 | Kidney | −8.9 | 3.15 | 0.61 | 1.1 | 0.56 | 3.08 | 1.31 | 5 | 347 | 0.05 | 1.3 | 0.68 | 0.05 | 3.28 | 0.4 | 63 | |
| F. attenuata | EC2006-02-02 | Muscle | −16.1 | 13.5 | <dl | <dl | 0.07 | <dl | <dl | 2 | 575 | 53.3 | <dl | <dl | 0.14 | <dl | <dl | 42 | |
| F. attenuata | EC2006-04-01 | Muscle | −15.9 | 12.0 | <dl | <dl | 0.04 | <dl | <dl | 2 | 621 | 56.8 | <dl | <dl | 0.69 | 16.5 | <dl | 29 | |
| F. attenuata | EC2006-04-02 | Muscle | −16.0 | 12.6 | <dl | <dl | 0.06 | <dl | <dl | 2 | 575 | 89.3 | <dl | <dl | 0.20 | 26.2 | <dl | 44 | |
| F. attenuata | EC2006-04-03 | Muscle | −16.0 | 12.7 | <dl | <dl | 0.03 | <dl | <dl | 3 | 531 | 13.9 | <dl | <dl | 0.23 | <dl | <dl | 43 | |
| F. attenuata | EC2006-07-01 | Muscle | −16.5 | 13.2 | <dl | <dl | 0.14 | <dl | <dl | 3 | 636 | 153 | <dl | <dl | <dl | 52.8 | <dl | 48 | |
| Mean | −16.1 | 12.8 | 0.07 | 2 | 587 | 73.3 | 0.32 | 31.9 | 41 | ||||||||||
| s.d. | 0.3 | 0.6 | 0.04 | 0 | 42 | 52 | 0.25 | 18.8 | 7 | ||||||||||
| G. macrorhynchus | EC1997-01-01 A | Muscle | −16.4 | 12.5 | ND | ND | 0.79 | <dl | <dl | 1 | 347 | 32.8 | <dl | <dl | ND | 8.56 | <dl | 61 | |
| G. macrorhynchus | EC1997-01-02 A | Muscle | −16.5 | 13.4 | ND | ND | 1.48 | <dl | <dl | 2 | 622 | 27.3 | <dl | <dl | ND | 5.62 | <dl | 50 | |
| G. macrorhynchus | EC2009-07-01 | Muscle | −15.7 | 11.2 | <dl | 2.6 | 0.85 | <dl | 0.74 | 2 | 653 | 9.93 | 0.3 | 0.16 | <dl | 3.40 | <0.38 | 61 | |
| G. macrorhynchus | EC2009-07-02 | Muscle | −15.6 | 11.1 | <dl | 3.2 | 2.62 | <dl | 1.01 | 2 | 804 | 14.0 | 0.4 | <dl | <dl | 4.79 | <0.35 | 55 | |
| G. macrorhynchus | EC2009-07-03 | Muscle | −16.0 | 11.0 | <dl | 3.8 | 0.56 | <dl | 0.47 | 2 | 664 | 9.94 | 0.3 | 0.21 | <dl | 3.56 | <0.4 | 51 | |
| Mean | −16.0 | 11.8 | 3.2 | 1.26 | 0.74 | 2 | 618 | 18.8 | 0.4 | 0.18 | 5.19 | 56 | |||||||
| s.d. | 0.4 | 1.1 | 0.6 | 0.83 | 0.27 | 1 | 167 | 10.6 | 0.0 | 0.03 | 2.10 | 5 | |||||||
| I. pacificus | EC2013-06-01 B | Muscle | −16.4 | 12.5 | <dl | 2.5 | 0.22 | <dl | 0.20 | 2 | 761 | 10.5 | 0.3 | <dl | <dl | 2.10 | <dl | 33 | |
| I. pacificus | EC2013-06-02 B | Muscle | −16.0 | 13.3 | <dl | 2.6 | 0.30 | <dl | 0.20 | 2 | 764 | 13.9 | 0.3 | <dl | <dl | 3.20 | <dl | 30 | |
| I. pacificus | EC2013-06-03 B | Muscle | −16.3 | 12.8 | <dl | 3.8 | 0.84 | <dl | 0.34 | 2 | 919 | 57.3 | 0.5 | <dl | <dl | 22.9 | <dl | 41 | |
| Mean | −16.2 | 12.8 | 2.9 | 0.45 | 0.25 | 2 | 815 | 27.2 | 0.4 | 9.4 | 35 | ||||||||
| s.d. | 0.2 | 0.4 | 0.7 | 0.34 | 0.08 | 0 | 90 | 26.1 | 0.1 | 11.7 | 6 | ||||||||
| K. breviceps | EC1997-04-01 A | Muscle | −15.7 | 13.2 | ND | ND | 0.41 | <dl | 0.03 | 2 | 977 | 6.07 | 0.3 | <dl | ND | 6.41 | <dl | 43 | |
| K. breviceps | EC1997-03-01 A | Muscle | −16.6 | 13.4 | ND | ND | 0.57 | <dl | <dl | 1 | 820 | 5.16 | 0.1 | <dl | ND | 2.49 | <dl | 67 | |
| K. breviceps | EC2007-05-01 | Muscle | −16.6 | 14.6 | <dl | <dl | 0.04 | 0.03 | 1.26 | 4 | 304 | 0.68 | <dl | <dl | <dl | <dl | <dl | 58 | |
| K. breviceps | EC2007-04-01 | Muscle | −16.5 | 12.4 | <dl | <dl | 0.13 | 0.04 | <dl | 2 | 691 | 4.91 | <dl | <dl | <dl | <dl | <dl | 39 | |
| K. breviceps | EC2007-03-01 | Muscle | −16.2 | 12.7 | <dl | <dl | 0.12 | 0.36 | 1.11 | 2 | 873 | 6.51 | 2.7 | <dl | <dl | <dl | <dl | 45 | |
| K. breviceps | EC2006-09-01 | Muscle | −17.0 | 12.8 | <dl | <dl | 0.21 | 0.03 | <dl | 2 | 746 | 7.73 | <dl | <dl | <dl | <dl | <dl | 51 | |
| K. breviceps | EC2010-04-01 | Muscle | −16.2 | 12.5 | <dl | <dl | 0.21 | 0.05 | 2.07 | 3 | 883 | 8.84 | 1.5 | <dl | <dl | 4.44 | <dl | 51 | |
| Mean | −16.4 | 13.1 | 0.24 | 0.10 | 1.12 | 2 | 756 | 5.70 | 1.2 | 4.45 | 51 | ||||||||
| s.d. | 0.4 | 0.8 | 0.19 | 0.14 | 0.84 | 1 | 221 | 2.61 | 1.2 | 1.96 | 10 | ||||||||
| K. sima | EC2011-01-01 | Muscle | −16.3 | 11.5 | <dl | 1.1 | <dl | 0.12 | 2.23 | 2 | 880 | 6.10 | 1.0 | 1.62 | 0.04 | 3.52 | <dl | 34 | |
| D. dugon | EC2006-01-01 | Muscle | −7.3 | 4.7 | <dl | <dl | 0.03 | 0.12 | <dl | 1 | 400 | 0.27 | <dl | <dl | 0.03 | <dl | <dl | 96 | |
| D. dugon | EC2006-13-01 | Muscle | −7.4 | 3.8 | <dl | <dl | 0.03 | 0.12 | <dl | 1 | 191 | 0.70 | <dl | <dl | 0.10 | <dl | <dl | 94 | |
| D. dugon | EC2009-01-01 | Muscle | −8.2 | 2.4 | <dl | <dl | <dl | 0.08 | <dl | 1 | 86 | 0.03 | <dl | <dl | 0.04 | <dl | <dl | 75 | |
| Mean | −7.6 | 3.6 | 0.03 | 0.11 | 1 | 226 | 0.33 | 0.05 | 88 | ||||||||||
| s.d. | 0.5 | 1.2 | 0.00 | 0.02 | 0 | 159 | 0.34 | 0.04 | 11 | ||||||||||
| G. macrorhynchus | EC1997-01-01 A | Blubber | ND | ND | ND | ND | 0.95 | <dl | 2.51 | 0.4 | 187 | 10.97 | 0.2 | <dl | ND | 4.44 | <dl | 21 | |
| G. macrorhynchus | EC1997-01-02 A | Blubber | ND | ND | ND | ND | 0.84 | <dl | 0.32 | 0.2 | 18 | 3.20 | <dl | <dl | ND | 3.02 | <dl | 17 | |
| D. dugon | EC2009-01-01 | Blubber | −5.2 | 3.4 | <dl | <dl | <dl | 0.23 | 0.54 | 0.3 | 12 | 0.01 | <dl | 0.21 | 0.06 | 0.12 | <dl | 9 | |
Wet:dry wt ratios are 4.5, 4.2, 4.0 and 1.6 for liver, kidneys, muscle and blubber, respectively.
dl, detection limit; ND, not determined.
Trace element concentrations also varied across both tissue type and species (Table 1). Specifically, muscle showed concentration values below 1 μg g−1 dry weight (dw) for Ag, As, Cd, Co, Cr, Mn, Ni, Pb and V, with a few exceptions (e.g. Cr and Ni in dwarf sperm whale). In contrast, Hg concentrations were high in the muscle of toothed whales (e.g. up to 153 μg g−1 dw in pygmy killer whales) but two to three times lower in dugong (between 0.68 and 8.84 μg g−1 dw). Similarly, trace elements in blubber were very low with the exception of Hg in toothed whales.
Liver and kidney showed high concentrations of several trace elements. Of these, the liver showed the highest concentrations of Ag, Cu, Fe, Hg, Se, and Zn and the kidney the highest for Cd. However, hepatic Hg concentrations were relatively low in dugongs (0.05–2.65 μg g−1 dw), had intermediate values in pygmy and dwarf sperm whales (1.15–50.5 μg g−1 dw) and had extremely high concentrations in Longman’s beaked whales (131–800 μg g−1 dw), pilot whales (1035–2290 μg g−1 dw) and pygmy killer whales (147–3959 μg g−1 dw). Se followed a similar pattern as Hg, with the lowest concentrations in the liver in dugongs and the highest in pygmy killer whales stranded in New Caledonia. Both Hg and Se were strongly correlated in the liver of toothed whales, indicating their coaccumulation in this tissue. A molar ratio between hepatic Hg and Se close to 1 was found in all specimens with the exception of the young individuals (calves and juveniles) for odontocetes and all dugongs whatever their age. It is remarkable that for pygmy killer whales, this molar ratio was also close to 1 in the kidneys and muscles. In dugongs specifically, very high concentrations of Co, Fe and Zn were found in the liver. The liver of marine mammals stranded in New Caledonia also contained high Cd concentrations but they remained below kidney levels.
Stable isotopes of carbon and nitrogen are widely used to infer the trophic ecology (δ13C for the feeding habitats and δ15N for the trophic position, (Hobson 1999)). In the marine mammals from New Caledonia, δ13C values in the different tissues reflect the segregation between inshore and offshore species. In each group, little difference between tissues indicates that the species exploit a relatively homogeneous habitat and that they do not make major changes in their habitat on both short (liver) and long (muscles) terms (Fig. S2). Values of δ15N clearly differentiate the herbivorous species (i.e. dugongs) from the toothed whales which, based on the trophic tracers, appear to have a relatively similar trophic niche. Nevertheless, when considering trace elements to infer the trophic ecology of toothed whales, the very high Cd concentrations in the liver and kidneys of short-finned pilot whales and Longman’s beaked whales suggest that they preferentially consume cephalopods (Bustamante et al. 1998). In a previous study, a large proportion of cephalopods (60% of the prey) was found in the stomachs of short-finned pilot whales stranded in New Caledonia (Bustamante et al. 2003). The very high concentrations of Hg in the tissues of toothed whales, particularly in the liver, suggest that they consume a large proportion of fish. It is remarkable that one individual pygmy killer whale had a hepatic Hg concentration close to 4000 μg g−1 dw. In toothed whales stranded in New Caledonia, the very high concentrations of Se in the liver, which correlate with that of Hg, reveals the existence of the demethylation of methyl-Hg (MeHg) by Se, which leads to a coaccumulation of the two elements in the form of mercuric selenide (HgSe) granules or tiemannite (Cuvin-Aralar and Furness 1991; Nigro and Leonzio 1996). Tiemannite granules have been reported in the liver of seabirds, marine mammals and humans (Martoja and Berry 1980; Pelletier 1986; Nigro and Leonzio 1996) but more recently in the kidneys, muscles and brains of giant petrels (Macronectes spp.) from the Southern Ocean (Manceau et al. 2021a) and the muscles of the blue marlin (Makaira sp., Manceau et al. 2021b). These results suggest that pygmy killer whale demethylates MeHg in the kidneys and muscles as well and raise the question of this detoxification process in their brains. Future studies on this species should be undertaken to answer this question.
Although the number of individuals presently studied remains modest, this study provides new information on poorly documented species (Longman’s beaked whale, pygmy killer whale, dwarf and pygmy sperm whales, dugong). Two studies already discussed trace elements in Longman’s beaked whales, short-finned pilot whales and pygmy sperm whales (Bustamante et al. 2003; Garrigue et al. 2016). Trace element concentrations in dugong tissues were generally similar to those previously reported in Australian dugongs (e.g. Denton et al. 1980; Denton and Breck 1981; Kemper et al. 1994). Strikingly, maximum concentrations of Co, Fe and Zn in the liver of dugongs are much higher than in the other species. The very high concentrations of Fe are related to its binding to ferritin, ferrihydrite and goethite iron oxide, resulting in reduced Fe toxicity (Rahman et al. 1999). As Co shares several similarities with Fe in its atomic properties (Stadler and Schweyen 2002; Rodrigue et al. 2005), it is possible that Co can be detoxified in the same way as Fe. Further research should be conducted to clarify this hypothesis. Considering the potential factors of variation of trace elements, the concentrations of Ag, Cd, Hg, and Se were higher in the older individuals compared with younger ones. Such a pattern is typical of marine mammals as a result of their bioaccumulation with age.
Focus on five taxa
In New Caledonia, the dugong is mainly found within the lagoons around the main island (Garrigue et al. 2008; Cleguer et al. 2015; Derville et al. 2022), therefore, sick or dying dugongs are more likely to be found in coastal areas. Dugong strandings occurred year-round with no significant seasonal effect (χ2 test χ2 = 2.5, P = 0.500), however, the number of stranding events have significantly increased since 1991 (z = 3.85, P < 0.001, deviance explained 0.33; Fig. 3a). Stranding events have mainly been observed on the west coast, where human and dugong densities are the highest, with two hotspots located around Nouméa and Ouano (Fig. 3c).
Dugong stranding data recorded since 1991. (a) Number of individuals stranded per year. (b) Number of individuals stranded per month. Stranding events are indicated with horizontal white lines to distinguish isolated and mass strandings based on number of individuals. (c) Geographical locations of events (with number of individuals represented as point size). (d) Causes of death including five individuals stranded alive with cause unknown and refloated.
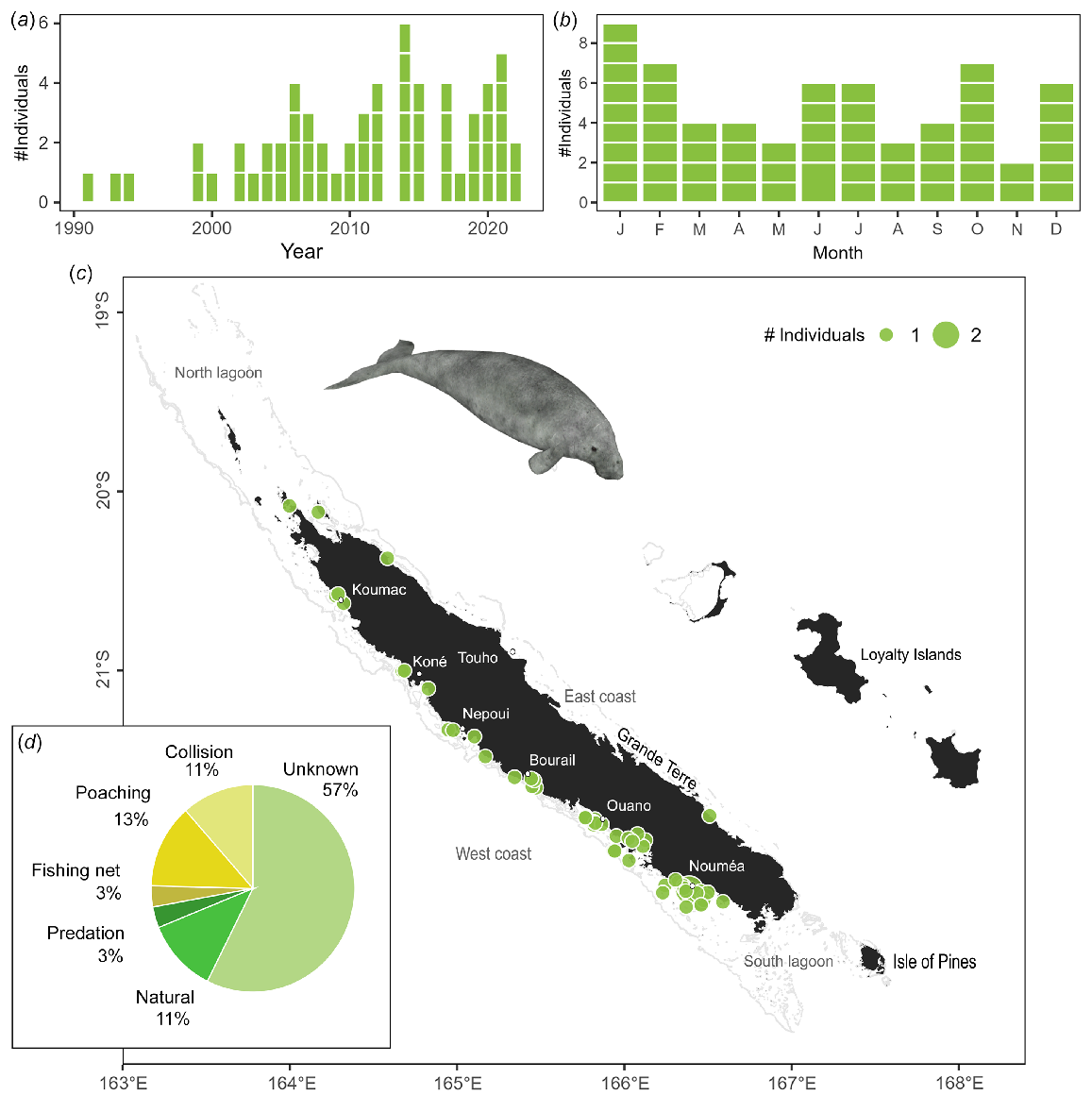
Sixty stranding events of solitary animals, including one stranding of a mother with a calf, were recorded (Table S3). Seven individuals were stranded alive and five successfully released, 48 stranded individuals varied from freshly dead (21% of them) to remains (19%), and no information was available on the state of the eight remaining carcasses.
Total length was obtained for 21 individuals, and ranged from 116 cm to 310 cm (Table S5). Following Lanyon et al. (2021), the mean total body length ± s.d. was calculated for calves (137.0 ± 30.6, n = 4, min = 116, max = 182), and adults (270.7 ± 19.1, n = 15). Adult females (279.5 cm ± 17.3, n = 6, min = 260, max = 301) were not significantly larger than adult males (264.9 cm ± 18.9, n = 9, min = 250, max = 310) on average (Welch t-test: t = 1.6, d.f. = 11.6, P = 0.134). The estimated age of eight individuals of which we could retrieve tusks for ageing ranged from 8 to 37 years.
Stomach content analysis of the species of plant material visually identified in the stomach of five dugongs confirmed a diet mainly composed of the seagrass species, Halophila ovalis, Halodule cf. uninervis, Syringodium isoetifolium, Cymodocea serrulata and Cymodocea sp., a diet previously identified by Marsh et al. (1982). Interestingly, each individual dugong stomach contained a different dominant seagrass species. Small fragments of seaweeds from Sargassum spp. and of Caulerpa spp. as well as some shells of Modiolus, fragments of sponges, and ascidians were also found in a 3.00 m-long female’s stomach (Brisset et al. 2022). Stable isotope analysis currently underway will allow further knowledge of the diet of dugongs in the area (Thibault et al. unpubl. data).
The cause of mortality was identified for almost half of the dugongs reported stranded between 1991 and 2022 (n = 26 of 61). Cause of death was determined to be of anthropogenic origin for 28% (n = 17) of cases (including illegal capture (poaching), boat collision, and incidental bycatch in fishing gear), of natural origin for 11% (n = 9) of cases (including diseases, meteorological events or predation) and could not be determined for the remaining 57% (n = 35, including five refloated animals) of cases (Fig. 3d). Seven cases of poaching were also found further inland but not included in analyses.
The dugong population of New Caledonia shows an extremely low mtDNA diversity, and a high genetic differentiation with other populations (Garrigue et al. 2022). This precarious genetic status was one of the key determinants that led the International Union for Conservation of Nature (IUCN) to revise the status of the New Caledonia dugong population, now listed as endangered (Hamel et al. 2022). The large number of deaths identified in this study from poaching or other anthropogenic causes is concerning. This strong anthropogenic pressure may weaken the population and endanger its survival, reinforcing the need for an urgent response from management and conservation institutions to protect this species in New Caledonia.
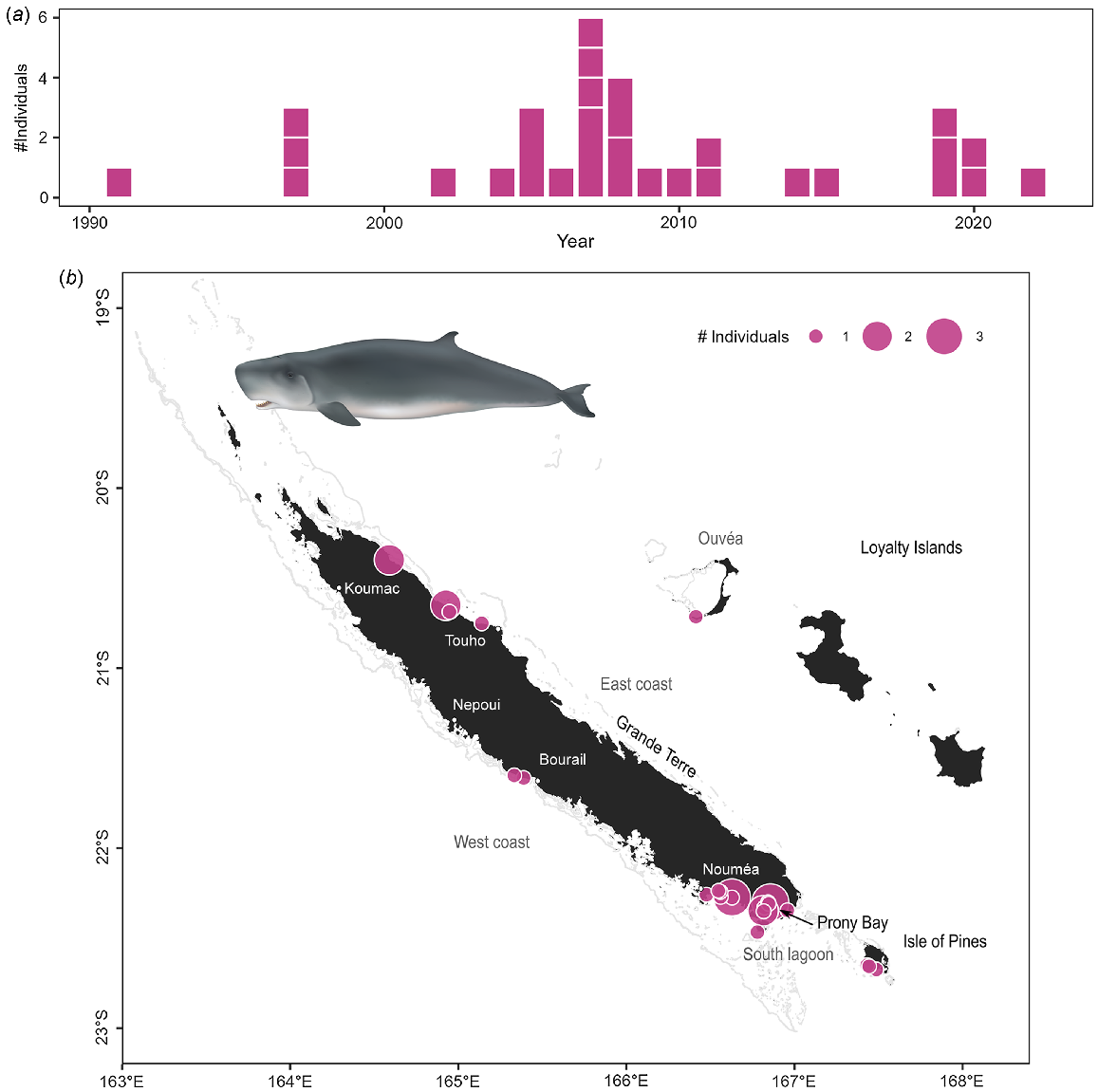
Sperm whales were the second most commonly stranded species in the archipelago (45 stranding events and 48 individuals Table S3), and the most common species to strand in the Loyalty Islands. Sperm whales stranded all around the archipelago, with some areas of higher concentrations: between Bourail and Nepoui on the west coast, around Touho on the east coast, and in the south lagoon including the Isle of Pines (Fig. 4c). In contrast to dugongs, there was no temporal trend observed in the number of stranding events of sperm whales since 1991 (Fig. 4a; z = 0.25, P = 0.799). Stranding events were documented year-round (Fig. 4b), consistent with reported year-round presence of the species in New Caledonian waters (Poupon and Garrigue 2011). There was no significant effect of season on the number of sperm whale strandings (χ2 test: χ2 = 3.00, P = 0.406) despite apparent peaks of events in May and November. Two stranding events involved newborn calves (total length <4 m Jefferson et al. 2008). Both of these events occurred toward the end of the austral summer (March–April) which, given a reported gestation period of 14–16 months in this species (Jefferson et al. 2008; Whitehead 2018), indicates mating occurred in the spring. Observations of large groups at sea during the spring (October–December pers. obs.) indicates that sperm whales may be both mating and calving in New Caledonian waters.
Stranding data recorded for sperm whales since 1877. Number of individuals stranded (a) per year and (b) per month. Stranding events are indicated with horizontal white lines to distinguish isolated and mass strandings based on number of individuals. (c) Geographical locations of events (with number of individuals represented as point size). Illustration credit: NOAA Fisheries.
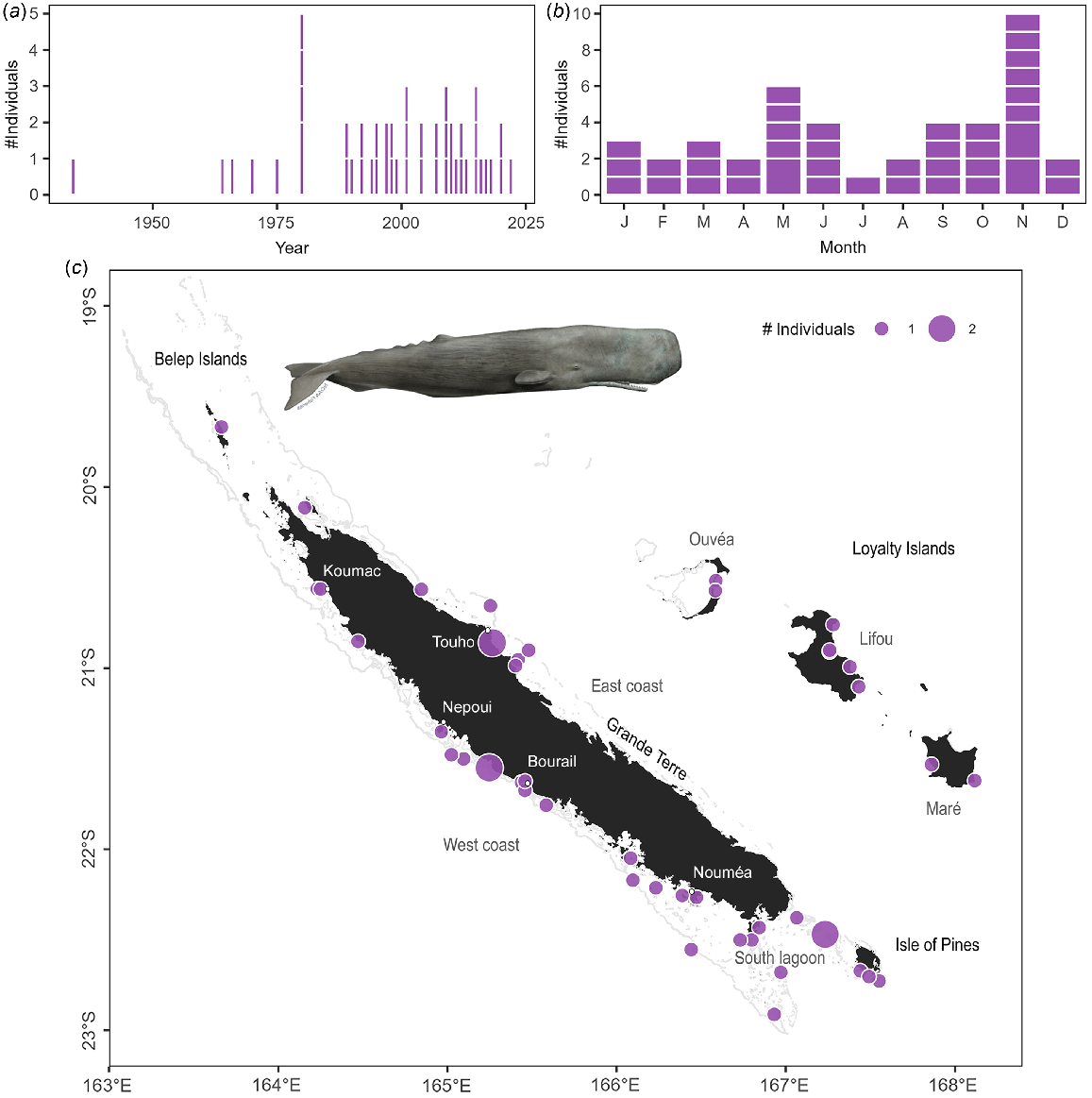
When carcasses were available for examination, they were partly decomposed and/or showed signs of having been scavenged at sea suggesting that the animals died at sea and were subsequently washed up on the reef. The head ‘case’ of sperm whales is composed of gristle, which is very tough and slow to decompose after death. Given the state of carcasses, few measurements or samples other than skin could be collected, and the species identification have subsequently been confirmed from DNA.
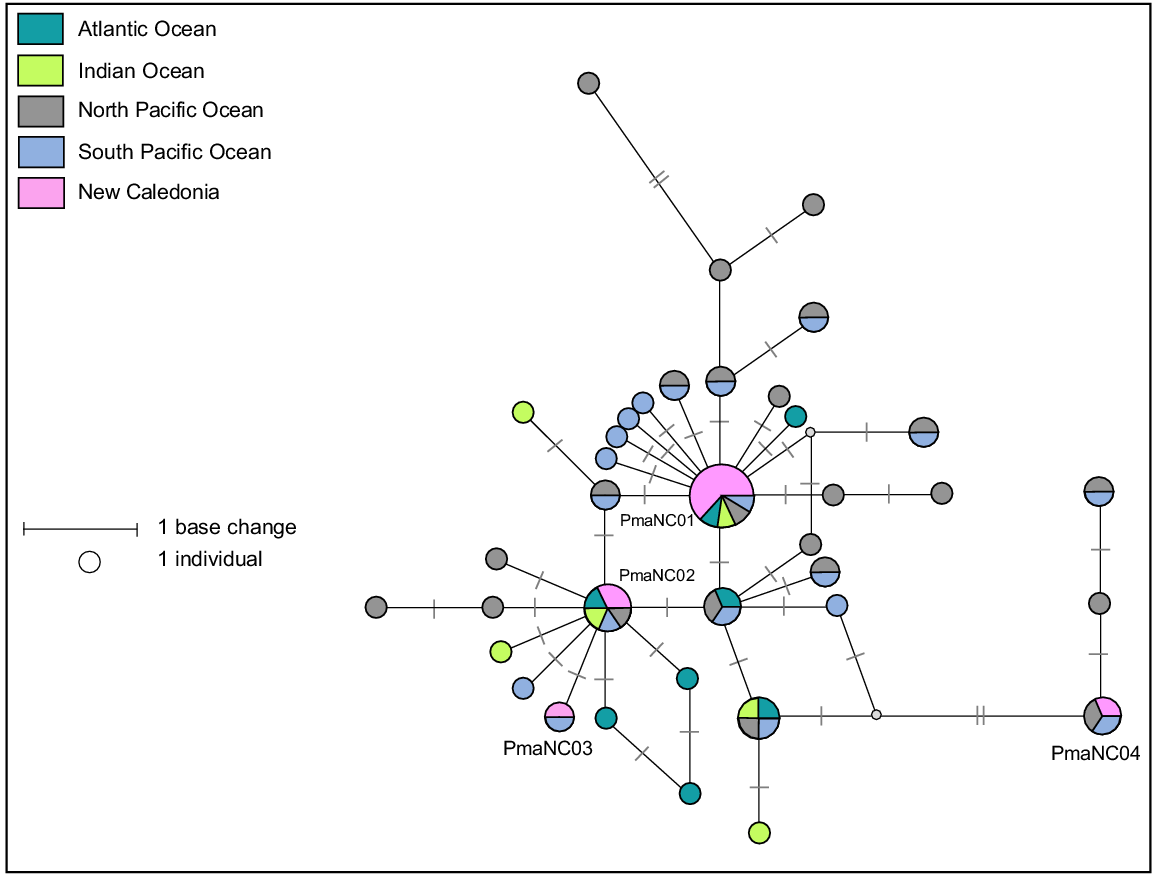
A 578-bp fragment of the mtDNA control region was successfully sequenced for eight samples collected from eight stranding events plus three biopsies samples taken from live animals in New Caledonia. Four haplotypes (Pma-NC01, Pma-NC02, Pma-NC03 and Pma-NC04), differing by seven base pairs, were identified (Fig. 5). Clustal W alignment sequences from the 11 samples and 222 others available were produced from a 301-bp consensus fragment of the mtDNA control region, containing 30 variable sites and leading to 37 different haplotypes that were already known from Alexander et al. (2016). The four New Caledonian haplotypes already known from previous studies correspond to haplotypes A, C, Z, O respectively (Alexander et al. 2016). The mtDNA control region haplotype diversity within New Caledonia sperm whales is Hd = 0.600, slightly lower than diversity estimates reported for other regions in the Pacific Ocean (ranging from 0.643 in Hawaii to 0.788 in the Gulf of California (Alexander et al. 2016)). Sperm whales are a long-lived species with few geographical barriers, however their worldwide mtDNA diversity is relatively low compared to other cetacean species with similar life-history traits (Lyrholm et al. 1996; Whitehead et al. 1998; Alexander et al. 2013, 2016). Although overall mtDNA diversity is low in this species, there exists a high genetic structure among geographic regions or social groups that is attributed to female philopatry (Lyrholm et al. 1999). In the Pacific Ocean, the importance of social group philopatry is strong and appears to be the main driver in genetic differentiation between regions (Alexander et al. 2016). Although genetic differentiation and fine-scale genetic structure was not tested here, the New Caledonian haplotypes found in strandings are consistent with those previously reported in the Pacific Ocean, indicating that is likely the basin of origin of these sperm whales.
Median-joining network based on a 302 bp alignment from 11 mtDNA control region sequences from sperm whales stranded in New Caledonia and 56 sperm whales worldwide reference haplotypes. Size of circle represents relative frequencies for New Caledonian sperm whales; colours represent putative populations of origin. The lengths of black lines represent the number of base changes. Except for individuals sampled in New Caledonia, only one sequence per haplotype per region has been represented here.
Species of the genus Kogia were the third most common animals to strand. The first record of a Kogia stranding is from 1972 and since 1991, 25 stranding events comprising 30 individuals have been reported. The stranding records, since 1991, show no temporal trend (Fig. 6a; z = 1.09, P = 0.276) nor any seasonal effect (χ2 test: χ2 = 1.67, P = 0.671). The majority of stranding events were of a single animal (n = 20), three were mother–calf pairs, and two were mass strandings involving three individuals. Nine individuals stranded alive in six events, and only one was successfully refloated. Kogia strandings were concentrated in the south and south-western part of the main island (67% of records since 1991), with a hotspot around Prony Bay (Fig. 6b, Table S3).
Stranding data recorded for dwarf and pygmy sperm whales since 1991. (a) Number of individuals stranded per year. Stranding events are indicated with horizontal white lines to distinguish isolated and mass strandings based on number of individuals. (b) Geographical locations of events (with number of individuals represented as point size).
Species identification (either from morphology or DNA analyses) was possible for 20 of the total 25 stranding events, five events of dwarf sperm whales (eight individuals of which five females, two males) and 15 events of pygmy sperm whales (17 individuals of which five females and five males).
Eight dwarf sperm whales and 14 pygmy sperm whales were in good enough condition for measurements and samples to be collected. For dwarf sperm whales, stranded adults ranged from 217 to 237 cm in length (Table S5) with a mean ± s.d. of 228.8 ± 7.9 cm (n = 5 individuals). The ratio between height of dorsal fin and total length ranged between 7.5 and 8.9%. Tooth age was estimated for five individuals, the youngest was 4.1, and the oldest was 15.7 years. Adult pygmy sperm whales ranged from 250 to 320 cm in length (Table S5) with a mean total length ± s.d. of 295.2 ± 26.4 cm (n = 10 individuals). The ratio between height of dorsal fin and total length ranged from 3.6 to 5.3%. Tooth age was estimated for six individuals and varied from 6.0 to 19.0 years.
A 504-bp fragment of the mtDNA control region was successfully sequenced for five samples of dwarf sperm whale collected from three stranding events. Four haplotypes (KsiNC01 to KsiNC04) differing at 15 bp were identified leading to a high haplotype diversity of Hd = 0.900. One of the events (EC2005-04) was a mass stranding of three individuals (two adult females and one male calf). Necropsy results of one of these adult females showed the presence of small ovaries (<10 cm) and a large womb as well as milk in the mammary glands, indicating a recent parturition. This female shared the same mtDNA haplotype (KsiNC01) with the stranded male calf suggesting this calf was her recent offspring.
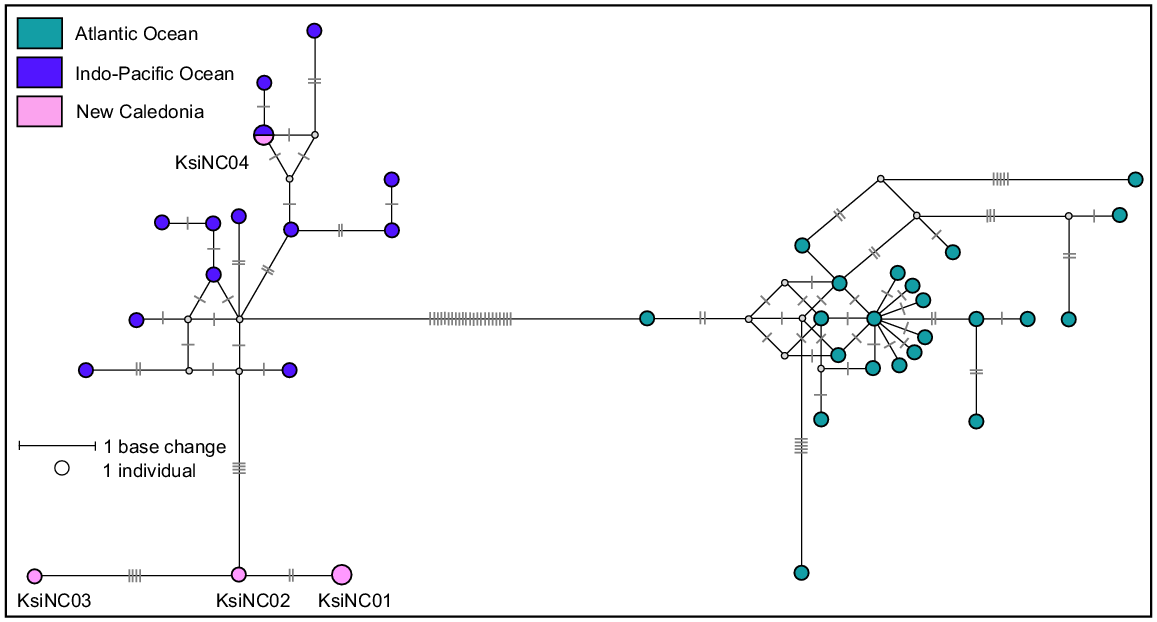
A total of 39 sequences previously published in Chivers et al. (2005) and Viricel (2012) were downloaded from GenBank and used as a reference dataset. After aligning the five sequences from this study, all sequences were trimmed to a 354 bp consensus length. This length describes 38 different haplotypes, separated into two distinct clades (Atlantic Ocean and Indo-Pacific Ocean), as previously reported in Chivers et al. (2005) and shown in the median-joining network (Fig. 7). The four haplotypes sequenced as part of this study all belong in the Indo-Pacific clade. One haplotype, KsiNC04, from a single stranded individual was identical to a previously published haplotype from the Indo-Pacific region (exact sample location unknown). The remaining three haplotypes (KsiNC01, KsiNC02 and KsiNC03) described in this study are new and separated from the rest of the Indo-Pacific clade by four base pairs. This is perhaps not surprising when one considers that of the 13 haplotypes represented in the Indo-Pacific clade reference dataset only one was from the Pacific Ocean (Chivers et al. 2005). Although dwarf sperm whales are widely distributed, they are elusive species that are difficult to approach at sea. For this reason, there are few opportunities to collect samples from living animals (Baird et al. 2022). The three new haplotypes described from stranded animals in this study are an important addition to knowledge of dwarf sperm whale genetic diversity in the Pacific Ocean.
Median-joining network based on a 354 bp alignment of five mtDNA control region sequences from dwarf sperm whales stranded in New Caledonia and 35 dwarf sperm whales worldwide reference haplotypes. Size of circle represents relative frequencies for New Caledonian dwarf sperm whales; colours represent putative populations of origin. The lengths of black lines represent the number of base changes. Except for individuals sampled in New Caledonia, only one sequence per haplotype per region has been represented here.
A 453-bp fragment of the mtDNA control region was successfully sequenced for two samples collected from pygmy sperm whales. These two sequences resolved two haplotypes that differed by 11 bp, KbrNC01 and KbrNC02. Neither of these haplotypes matched previously identified haplotypes from the Indo-Pacific Ocean (n = 29) or the Atlantic Ocean (n = 67) (Chivers et al. 2005; Viricel 2012). In contrast to dwarf sperm whales, there is no evidence of global geographic structure in pygmy sperm whales (Chivers et al. 2005). A haplotype network constructed from the New Caledonia sequences and the reference dataset confirmed this lack of geographic structure and showed both haplotypes from New Caledonia nested in a global haplotypic network (network not shown).
No clear evidence for the cause of death was discovered but some individuals were clearly heavily infected by parasites. Anisakis physeteris and Anisakis paggiae (both Nematoda, Anisakidae), Phyllobothrium delphini and Monorygma grimaldii (both Cestoda, Phyllobothriidae) were respectively found in stomachs, blubber and muscles of pygmy sperm whale (Supplementary Material 2). Stomach nematodes have been commonly reported in stranded specimens of Kogia and several authors have suggested that individuals with parasites could suffer by expending energy feeding parasites rather than themselves. However, there is no quantitative study on stomach parasites that can report a threshold of infection leading to strandings (Mcalpine et al. 1997; Bustamante et al. 2003; Plön 2004).
Ten stomach contents were analysed. Two of them have been previously reported (Bustamante et al. 2003); two others (dwarf sperm whale EC2011-01-01 and pygmy sperm whale EC2019-07-01) were empty or contained an unrecorded quantity of parasites; results for the remaining six stomach contents are reported here (pygmy sperm whale n = 3 and dwarf sperm whale n = 3). Of the six stomach contents analysed, three were in a very advanced state of digestion (dwarf sperm whale EC2005-04-02 and EC2005-04-03 and pygmy sperm whale EC2007-03-01) and identifications were limited to less precise taxonomic level (e.g. Decapodiformes, Crustacea, Actinopterygii). There were a total of 599 prey items analysed from the six stomachs, from which 17 taxa were identified (Table S6). Information about the total number and the total weight of prey by stomach are summarised in Table 2. Proportions of prey items by number (%N), by weight (%W) and frequencies of occurrence (%FO) for these six stomachs are presented in Table S6. Cephalopods were present in 100% of the stomachs analysed but the majority of them were completely digested and only represented by beak remains that have not been identified but preserved in 80% ethanol for further identification (1 Octopodiforme, 0.02 g, and 575 Decapodiformes; 130.57 g). As cephalopod’ beaks could remain in the stomach for a long time, accumulating until they are regurgitated (Clark and MacLeod 1982; Staudinger et al. 2014), and to limit biases in the results, these hard parts have been omitted from the quantitative analysis. However, cephalopods still dominated the stomach contents by weight (58.9 %W). Among the remaining cephalopods, four were all partially digested (298.1 g) and were identified as one Histioteuthidae (4.35 %N, 1.44 %W), one Ommastrephidae (4.35 %N, 32 %W), one Ornithoteuthis sp. (4.35 %N, 5.35 %W) and one Histioteuthis meleagroteuthis (4.35 %N, 6.29 %W). Crustaceans were the second most important prey category of the Kogia’s diet (29.44 %W), with remains found in five of the six stomachs (83.33 %FO). Three families of deep-water shrimp were identified (Oplophoridae, Pasiphaeidae and Gnathophausiidae). Remains of the family Pasiphaeidae were found in two of the six stomachs (>4.35 %N, 1.72 %W), remains of the shrimp genus Gnathophausia sp. were found in one stomach (30.4 %N, 2.01 %W), and one shrimp of the genus Oplophorus was found in one stomach (4.35 %N, 0.23 %W). Fish also contributed to Kogia’s diet (66.67 %FO, 11.25 %W). Among the six stomachs analysed, four contained fish remains that could not be identified beyond class (all Actinopterygii) and one contained a fish of the family Tetraodontidae otherwise known as pufferfish (4.35 %N, 3.66 %W). Nematodes and other worm-like parasites that could not be identified were found in all six stomachs.
| Taxa | Code of individual | Total number of items | Total weight of items (g) | Total number of taxa | Stomach content examiner | Type of analysis | |
|---|---|---|---|---|---|---|---|
| K. breviceps | EC1997-04-01A | ND | ND | ND | IRD, MNHN | Whole content, morphological analysis, cephalopod beak identification | |
| EC1997-03-01A | ND | ND | ND | ||||
| EC2007-03-01 | 79 | 34.03 | 3 | SPC | Whole content, morphological analysis | ||
| EC2007-04-01 | 303 | 462.99 | 9 | ||||
| EC2010-04-01 | 63 | 101 | 3 | ||||
| EC2019-07-01 | Empty | IRD | In situ analysis | ||||
| K. sima | EC2004-01-01 | 109 | 183.49 | 9 | SPC | Whole content, morphological analysis | |
| EC2005-04-02 | 23 | 7.6 | 2 | ||||
| EC2005-04-03 | 22 | 2.6 | 4 | ||||
| EC2011-01-01 | Empty | IRD | In situ analysis | ||||
| Total | 599 | 791.71 | 17 |
IRD, French National Institute for Sustainable Development; MNHN, National Museum of Natural History (Paris, France); SPC, Pacific Community, Noumea, New Caledonia.
The analysis of stomach contents presented here indicates that, in this region, both species of Kogia feed mainly on squid, and complete their diet with crustaceans and a few fish. These results are consistent with those reported previously from New Caledonia strandings by Bustamante et al. (2003) and those reported by West et al. (2009) for the diet of pygmy sperm whales in the Hawaiian Archipelago. For both species of Kogia, the prey items identified do not all belong to one layer in the ocean, but rather span the epipelagic, mesopelagic and bathypelagic layers (West et al. 2009; cephalopods see Young and Vecchione 2009, crustaceans see Meland and Aas 2013, fish see Roberts et al. 2015). In some cases, identified prey items are also known to carry out diel vertical migrations (Histioteuthis meleagroteuthis, (Quetglas et al. 2010); Pasiphaea, (Cartes 1993)). This distribution of prey in the water column makes it difficult to estimate the depth at which either species of Kogia is feeding. Previous studies of diet suggest that pygmy sperm whales generally feed in deep shelf and slope waters (Santos et al. 2006). Clarke (2003) suggested that both species of Kogia could dive between 500 and 1000 m because they share the same prey species as sperm whale. Some authors believe the pygmy sperm whale takes its prey at or near the bottom because of the presence of benthic fishes and crabs and also because of its ‘small underslung lower jaw and anterio-ventrally flattened snout’ (Santos et al. 2006).
Kogia are cryptic species and their elusive behaviour make them difficult to visually observe at sea. To date, only one visual observation of a pygmy sperm whale in the Loyalty islands was confirmed. Nevertheless, the number of strandings of live animals (28% of the stranded Kogia) suggests that these species may be more common in New Caledonian waters than the low number of observations at sea suggests. A recent study using a passive acoustic monitoring method highlighted that visual surveys underestimate the presence of Kogia (Hildebrand et al. 2019).
More than 70% of the stranding events were found in the south-west part of the main island and more than half of those occurred between October and March. Hénin and Cresswell (2005) identified an upwelling of cold water, about 10 km wide, outside the western barrier reef, more commonly observed in summer than in winter (October to March) and related to south-easterly wind events. This oceanographic feature is likely to result in relatively more productive waters that could attract Kogia, especially as this teuthophageous feeding species is known to associate with dynamic frontal zones and eddies (Virgili et al. 2018).
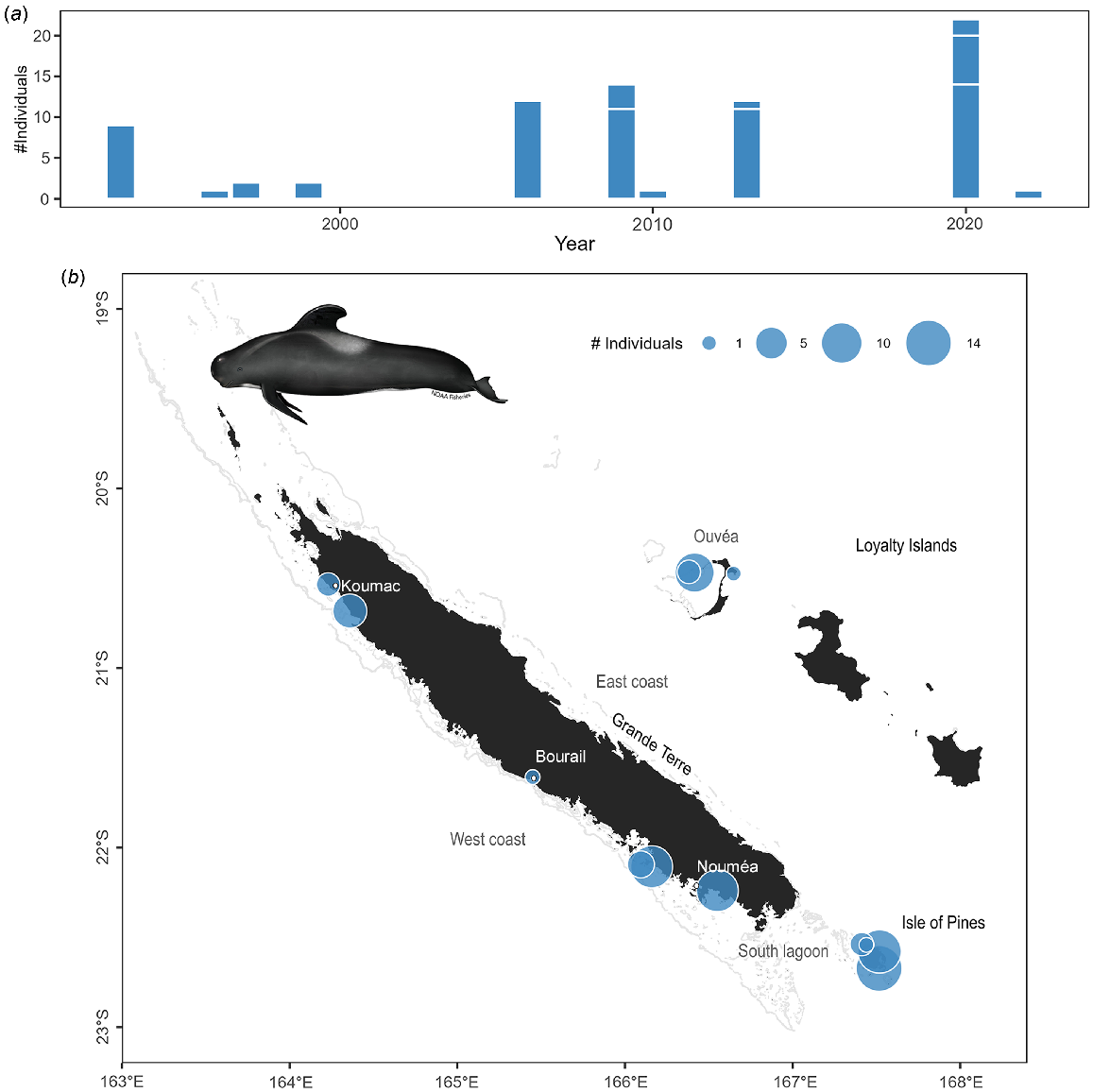
Although pilot whales account for only 7% of the stranding events (n = 16), they represent 35% (n = 127) of stranded individuals due to their tendency to mass strand (42% of the mass strandings reported). The 11 mass strandings documented in New Caledonia involved 2–50 individuals, but most of these events were of 10–15 animals. When interventions were possible most of the stranded animals were successfully refloated by rangers or the public.
There was no temporal trend observed in the number of stranding events of pilot whales (Fig. 8a; z = 1.03, P = 0.301), nor significant seasonal differences (χ2 test: χ2 = 2.1, P = 0.623) since more systematic record collection beginning in 1991. The majority of stranding events occurred in the south-western part of the main island and in the Isle of Pines (Fig. 8b).
Stranding data recorded for short-finned pilot whales since 1991. (a) Number of individuals stranded per year. Stranding events are indicated with horizontal white lines to distinguish isolated and mass strandings based on number of individuals. (b) Geographical locations of events (with number of individuals represented as point size). Illustration credit: NOAA Fisheries.
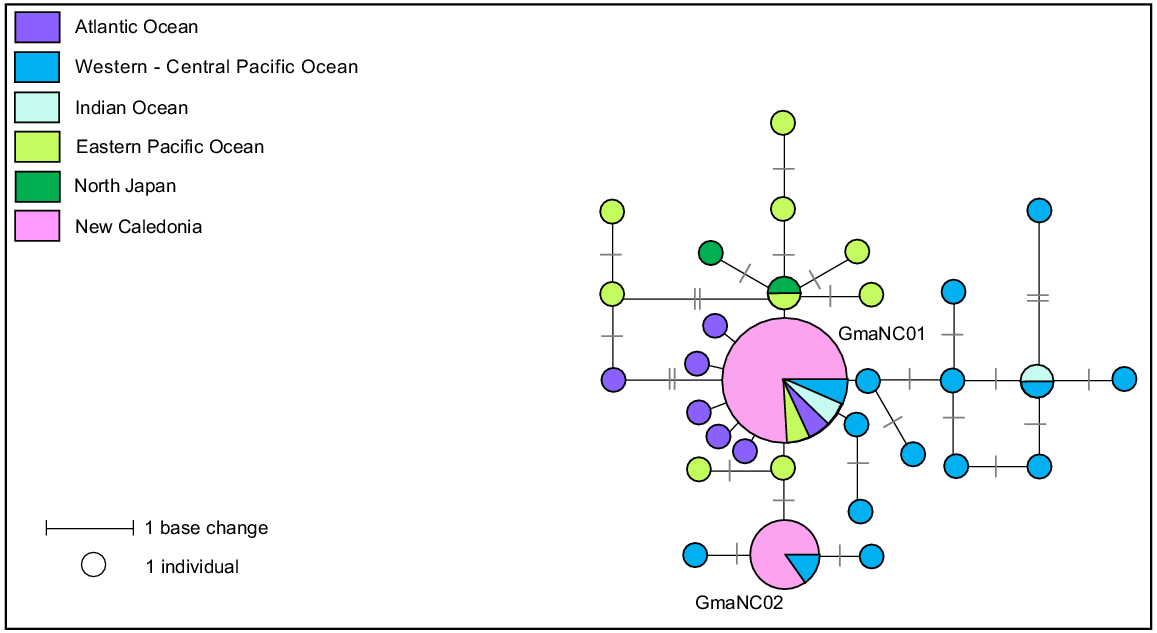
A 660-bp fragment of the mtDNA control region was successfully sequenced for 17 samples collected in New Caledonia (12 females and five males) from six stranding events. Two haplotypes that differed by two base pairs were identified, GmaNC01 and GmaNC02 (Hd = 0.309).
Fourteen of the 17 stranded individuals shared haplotype GmaNC01, and the remaining three individuals shared haplotype GmaNC02. Both haplotypes were found in three of the seven mass-stranding events for which samples were available. The social structure of both species of pilot whales has been described as matrilineal, with several generations of maternally-related individuals associating in the same group (Amos et al. 1993; Heimlich-Boran 1993). Multiple previous studies of pilot whales have shown that small groups are likely made up of related individuals, suggesting some degree of matrilineal philopatry, whereas large groups are probably temporary associations of these smaller groups (short-finned pilot whales – Madeira (Alves et al. 2013), Mariana Archipelago (Hill et al. 2019), Hawaii (Mahaffy et al. 2015); long-finned pilot whales – New Zealand (Oremus et al. 2013), Australia (Mahaffy et al. 2015; Hill et al. 2019)). This temporary formation of larger groups may be explained by temporary associations of social groups during mating seasons (Hill et al. 2019) or interactions between unrelated social groups during feeding (Oremus et al. 2013). The three mixed haplotype mass-stranding events (Table S5) around New Caledonia suggest short-finned pilot whales in this region also form larger aggregations of unrelated matrilines. Because calving is diffusely seasonal for the southern form with a peak in June–August but could happen year-round (Kasuya 2017), these three events that occurred in three different periods (February, May and September) could be linked to mating aggregations.
A total of 112 sequences were downloaded from GenBank to form a geographic reference dataset for this species. This dataset was then aligned with the 17 sequences generated in this study and trimmed to a consensus length of 335-bp. The reduced sequence length described a total of 31 haplotypes, defined by 25 variable sites (median-joining network, Fig. 9). Previous studies highlighted that the short-finned pilot whale includes at least two distinct morphological forms, the ‘Naisa’ and ‘Shiho’ forms (Yamase 1760). These morphological forms have been examined at the molecular level based on mitogenomes and nuclear single nucleotide polymorphism (SNP) loci (Van Cise et al. 2019). Based on mitogenome, authors suggested the existence of four genetic clades within the species: the clade ‘Atlantic’, the clade ‘Shiho’ encountered on the eastern Pacific Ocean and northern Japan, the clade ‘Naisa’ encountered on the western/central Pacific and Indian Ocean, and the ‘Clade 3’ having the same distribution of the ‘clade Naisa’ extending into the eastern Pacific Ocean. As for other regions in the South Pacific, both mitogenomes clades ‘Nasia’ and ‘Clade 3’ are represented in New Caledonia, as they matched with GmaNC01 and GmaNC02 respectively.
Median-joining network based on a 335 bp alignment of 17 mtDNA control region sequences from short-finned pilot whales stranded in New Caledonia and 31 short-finned pilot whales worldwide reference haplotypes. Size of circles represents relative frequencies for New Caledonian pilot whales; colours represent putative populations of origin. The lengths of black lines represent the number of base changes. Except for individuals sampled in New Caledonia, only one sequence per haplotype per region has been represented here.
Striped dolphins were first identified in New Caledonia during a mass-stranding event that occurred in the south lagoon, in the western part of Prony Bay, on 18 August 2019. A total of 11 animals stranded, seven were dead before help arrived, two were successfully rescued, and two died during the rescue attempt.
A 454-bp fragment of the mtDNA control region was successfully sequenced for eight individuals from this mass-stranding event. This fragment length resolved seven haplotypes with 28 variable sites. Only two (one male, one female) of the eight individuals shared the same mtDNA haplotype and, unlike the putative dwarf sperm whale cow–calf pair, there is no physical evidence to suggest these two form a parent–offspring pair.
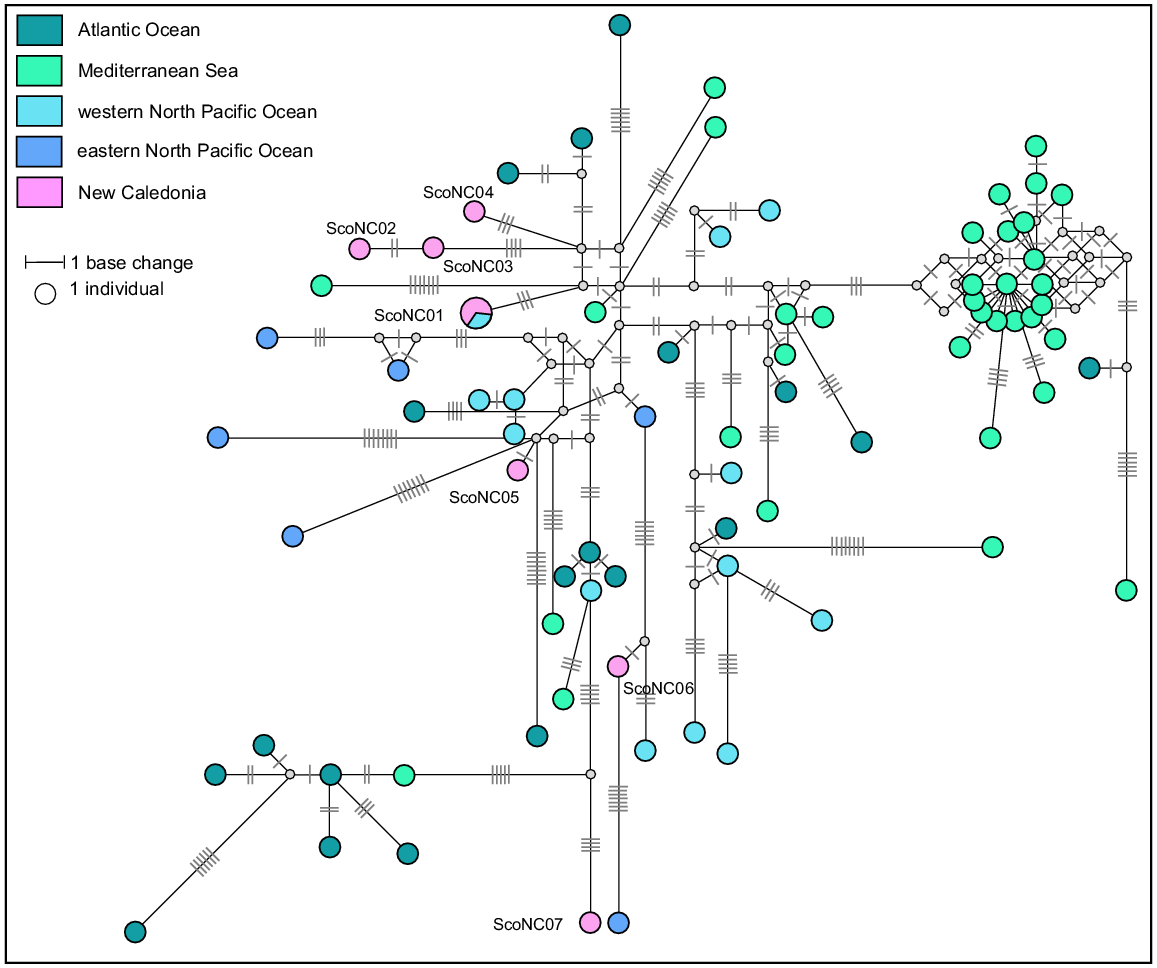
A total of 107 sequences were downloaded from GenBank to form a geographic reference dataset for striped dolphins. These were aligned with the eight sequences identified in New Caledonia and trimmed to provide a final dataset of 75 haplotypes (median-joining network, Fig. 10). There was only one shared haplotype between New Caledonia and the global reference dataset, ScoNC01. This haplotype was identified in two individuals stranded in New Caledonia and in one individual from the China Seas, and is the only haplotype shared between any regions in this dataset. No phylogeographic signal was detected in the network, however there is a lack of available published data from the South Pacific for comprehensive comparison of New Caledonia with other locations in this region.
Median-joining network based on a 458 bp alignment of eight mtDNA control region sequences from striped dolphins stranded in New Caledonia and 75 striped dolphins worldwide reference haplotypes. Size of circles represents relative frequencies for New Caledonian striped dolphins; colours represent putative populations of origin. The lengths of black lines represent the number of base changes. White dots represent unsampled median haplotypes. Except for individuals sampled in New Caledonia, only one sequence per haplotype per region has been represented here.
The results highlighted here are consistent with those previously published on the genus Stenella (Faria et al. 2022). For many species of Stenella, genetic diversity is high compared to other Delphinidae species (e.g. Caballero et al. (2013)) and no strong phylogeographic signal is detected.
Conclusions and perspectives
The monitoring of stranding events in New Caledonia over several decades has allowed for the collection of a great variety of biological and ecological data, contributing to an increased knowledge of marine mammal species in this region, which can help guide conservation measures. This surveillance was made possible through collaboration between scientists, governmental and provincial services, and the public, including volunteer veterinarians. Since 2016, training and outreach were provided to rangers, veterinarians, and various public safety officers to support their engagement in the scientific monitoring of stranding events. In support of this rudimentary, yet efficient, stranding network, a stranding monitoring website (www.rescue.ird.nc) was developed (Wahoulo 2019) and has been maintained online by IRD Nouméa since 2018. As of today, the objectives of this repository and platform are to (1) allow the stranding network field agents to report data in a standardised format; (2) create a centralised, online, and secure database for managers and scientists to retrieve information; (3) generate annual stranding listings to communicate to the French National Stranding Network (https://www.observatoire-pelagis.cnrs.fr/echouages/reseau-national-echouage/), and (4) provide a publicly accessible interactive map, displaying the locations of all marine mammal strandings in New Caledonia. In addition to being a great source of scientific knowledge, strandings are also exceptional and thought-provoking events that stimulate the public’s awareness of marine mammal protection and ocean conservation at large. For these reasons, the New Caledonian long-term stranding monitoring acts as a reference for the South Pacific and as such, warrants continued support going forward.
Data availability
The data that support this study are available in the article and accompanying online supplementary material as well as online: www.rescue.ird.nc. All details of the stranding events are available for each administrative entities respectively for the Provinces Nord, des Iles and Sud at https://doi.org/10.23708/VJCSYH, https://doi.org/10.23708/DVJ1YT, https://doi.org/10.23708/GPYMHX.
Declaration of funding
Funding for diet analysis was provided by the European Union ‘Pacific-European-Union-Marine-Partnership’ Programme (agreementFED/2018/397-941).
Author contributions
Conceptualisation (CG, SD), data collection (CG, CC, EC, PP, JCV), data curation (CG, PB, SF, JLJ, PM, AP, EV), formal analysis (CG, SD, CB, MB, PB, CC, EC, WD, JLJ, PM, TM, PP, AP, CS, DS, JCV, EV), funding acquisition (EV), project administration (CG), software (SF), visualisation (SD, CB, PB), writing (CG, SD, CB, PB, CC, EC, WD, SF, PM, AP, DS, EV)
Acknowledgements
This article is dedicated to the memory of our colleague and friend J C Vivier, one of the first veterinarians to be interested in stranded animals in NC and who tragically disappeared. We would like to thank all the people who reported strandings as well as those who collected data. We would like to especially thank the rangers of the northern, southern and islands provinces, as well as the veterinarians (Yann Baron, Sylvana Bima, Agathe Binois, Alexandra Campos, Anais Creff, Sophie Dinkel, Hervé Leroux, Sarah Vandeweeghe, Sarah Wund) who actively participated in monitoring the strandings. Genetic analysis was conducted at the Laboratory of Molecular Ecology and Evolution, University of Auckland, New Zealand and at the Plateforme du Vivant (IRD, Noumea, New Caledonia). The authors are grateful to Carine Churlaud and Maud Brault-Favrou from the Analyses Elémentaires platform (LIENSs) for their support during trace element analysis and to Gaël Guillou from the Analyses Isotopiques platform (LIENSs) for running the stable isotope analyses. Thanks are due to the CPER (Contrat de Projet Etat-Région) and the FEDER (Fonds Européen de Développement Régional) for funding the AMA, the ICP and the IRMS of LIENSs laboratory. Paco Bustamante is an honorary member of the IUF (Institut Universitaire de France). Thanks to the scientists who identified the parasites and especially to Professor Louis Euzet (University of Montpellier, France), Professor Alain Crosnier, Dr Annie Petter, and Professor Ian Beveridge from Australia.
References
Alexander A, Steel D, Slikas B, Hoekzema K, Carraher C, Parks M, Cronn R, Baker CS (2013) Low diversity in the mitogenome of sperm whales revealed by next-generation sequencing. Genome Biology and Evolution 5, 113-129.
| Crossref | Google Scholar | PubMed |
Alexander A, Steel D, Hoekzema K, Mesnick SL, Engelhaupt D, Kerr I, Payne R, Baker CS (2016) What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Molecular Ecology 25, 2754-2772.
| Crossref | Google Scholar | PubMed |
Allain V, Menkes C (2011) Nectalis 1-5 cruises. RV Alis. Available at https://doi.org/10.18142/243
Allain V, Menkes C (2021) Warmalis1 cruise. RV Alis. Available at https://doi.org/10.17600/18000710
Alves F, Quérouil S, Dinis A, Nicolau C, Ribeiro C, Freitas L, Kaufmann M, Fortuna C (2013) Population structure of short-finned pilot whales in the oceanic archipelago of Madeira based on photo-identification and genetic analyses: implications for conservation. Aquatic Conservation: Marine and Freshwater Ecosystems 23, 758-776.
| Crossref | Google Scholar |
Amos B, Schlotterer C, Tautz D (1993) Social structure of pilot whales revealed by analytical DNA profiling. Science 260, 670-672.
| Crossref | Google Scholar | PubMed |
Andréfouët S, Chagnaud N, Chauvin C, Kranenburg CJ (2008) Atlas of French Overseas coral reefs. Nouméa, New Caledonia. Available at http://umr-entropie.ird.nc/index.php/home/resources
Baird RW, Mahaffy SD, Lerma JK (2022) Site fidelity, spatial use, and behavior of dwarf sperm whales in Hawaiian waters: using small-boat surveys, photo-identification, and unmanned aerial systems to study a difficult-to-study species. Marine Mammal Science 38, 326-348.
| Crossref | Google Scholar |
Beasley I, Cherel Y, Robinson S, Betty E, Gales R (2013) Pygmy sperm whale (Kogin breiJiceps) stranding record in Tasmania, Australia, and diet of a single specimen. Papers and Proceedings of the Royal Society of Tasmania 147, 25-31.
| Crossref | Google Scholar |
Bonneville CD, Derville S, Luksenburg JA, Oremus M, Garrigue C (2021) Social structure, habitat use and injuries of Indo-Pacific Bottlenose Dolphins (Tursiops aduncus) reveal isolated, coastal, and threatened communities in the South Pacific. Frontiers in Marine Science 8, 606975.
| Crossref | Google Scholar |
Borsa P (2006) Marine mammal strandings in the New Caledonia region, Southwest Pacific. Comptes Rendus Biologies 329, 277-288.
| Crossref | Google Scholar |
Borsa P (2022) Présence saisonnière de l ’ otarie à fourrure dans la partie orientale de la mer de Corail. Bulletin de la Société Zoologique de France 147, 57-60 hal-03698720.
| Google Scholar |
Borsa P, Hoarau G (2004) A pygmy blue whale (Cetacea: Balaenopteridae) in the inshore waters of New Caledonia. Pacific Science 58, 579-584.
| Google Scholar | PubMed |
Brisset M, Derville S, Andréfouët S, Cleguer C, Buttin J, Garrigue C (2022) Science en herbe. Raising awareness about the importance of dugongs and their habitat in New Caledonia. Final report. French National Research Institute for Sustainable Development, Nouméa, New Caledonia. 26p + annexes. Available at https://hal.science/hal-04247094
Bustamante P, Caurant F, Fowler SW, Miramand P (1998) Cephalopods as a vector for the transfer of cadmium to top marine predators in the north-east Atlantic Ocean. Science of The Total Environment 220, 71-80.
| Crossref | Google Scholar | PubMed |
Bustamante P, Garrigue C, Greaves J, Dodemont R, Dabin W, Breau L (2001) Heavy metals contents in two odontocetes (Globicephala macrorhynchus and Kogia breviceps) stranded in New Caledonia (South Pacific). In ‘XIVth Biennial Conference on the Biology of Marine Mammals’. (Society for Marine Mammalogy: Vancouver, Canada)
Bustamante P, Garrigue C, Breau L, Caurant F, Dabin W, Greaves J, Dodemont R (2003) Trace elements in two odontocete species (Kogia breviceps and Globicephala macrorhynchus) stranded in New Caledonia (South Pacific). Environmental Pollution 124, 263-271.
| Crossref | Google Scholar | PubMed |
Caballero S, Marcos MCdO, Sanches A, Mignucci-Giannoni AA (2013) Initial description of the phylogeography, population structure and genetic diversity of Atlantic spotted dolphins from Brazil and the Caribbean, inferred from analyses of mitochondrial and nuclear DNA. Biochemical Systematics and Ecology 48, 263-270.
| Crossref | Google Scholar |
Cartes JE (1993) Feeding habits of pasiphaeid shrimps close to the bottom on the Western Mediterranean slope. Marine Biology 117, 459-468.
| Crossref | Google Scholar |
Chivers SJ, Leduc RG, Robertson KM, Barros NB, Dizon AE (2005) Genetic variation of Kogia spp. with preliminary evidence for two species of Kogia sima. Marine Mammal Science 21, 619-634.
| Crossref | Google Scholar |
Clark MR, MacLeod N (1982) Cephalopod remains from the stomachs of Sperm Whales caught in the Tasman Sea. Memoirs of the National Museum of Victoria 43, 25-42.
| Crossref | Google Scholar |
Clarke M (2003) Production and control of sound by the small sperm whales, Kogia breviceps and K. sima and their implications for other Cetacea. Journal of the Marine Biological Association of the United Kingdom 83(2), 241-263.
| Crossref | Google Scholar |
Cleguer C, Grech A, Garrigue C, Marsh H (2015) Spatial mismatch between marine protected areas and dugongs in New Caledonia. Biological Conservation 184, 154-162.
| Crossref | Google Scholar |
Cleguer C, Garrigue C, Fuentes MMPB, Everingham Y, Hagihara R, Hamann M, Payri C, Marsh H (2017) Drivers of change in the relative abundance of dugongs in New Caledonia. Wildlife Research 44, 365-376.
| Crossref | Google Scholar |
Clua E (2002) Présence et mort d’une baleine bleue sur les côtes néo-calédoniennes: quels enseignements scientifiques en tirer? Bulletin du Groupement Technique Vétérinaire 28, 25-29.
| Google Scholar |
Clua EE, Manire CA, Garrigue C (2014) Biological data of pygmy killer whale (Feresa attenuata) from a mass stranding in New Caledonia (South Pacific) associated with Hurricane Jim in 2006. Aquatic Mammals 40, 162-172.
| Crossref | Google Scholar |
Cox TM, Ragen TJ, Read AJ, Vos E, Baird RW, Balcomb K, Barlow J, Caldwell J, Cranford T, Crum L, et al. (2006) Understanding the impacts of anthropogenic sound on beaked whales. Journal of Cetacean Research and Management 7, 177-187.
| Crossref | Google Scholar |
Cuvin-Aralar MLA, Furness RW (1991) Mercury and selenium interaction: a review. Ecotoxicology and Environmental Safety 21, 348-364.
| Crossref | Google Scholar | PubMed |
Dalebout ML, Robertson KM, Frantzis A, Engelhaupt D, Mignucci-Giannoni AA, Rosario-Delestre RJ, Baker CS (2005) Worldwide structure of mtDNA diversity among Cuvier’s beaked whales (Ziphius cavirostris): implications for threatened populations. Molecular Ecology 14, 3353-3371.
| Crossref | Google Scholar | PubMed |
Denton GRW, Breck WG (1981) Mercury in tropical marine organisms from north Queensland. Marine Pollution Bulletin 12, 116-121.
| Crossref | Google Scholar |
Denton GRW, Marsh H, Heinsohn GE, Burdon-Jones C (1980) The unusual metal status of the dugong Dugong dugon. Marine Biology 57, 201-219.
| Crossref | Google Scholar |
Derville S, Torres LG, Dodémont R, Perard V, Garrigue C (2019) From land and sea, long-term data reveal persistent humpback whale (Megaptera novaeangliae) breeding habitat in New Caledonia. Aquatic Conservation: Marine and Freshwater Ecosystems 29, 1697-1711.
| Crossref | Google Scholar |
Derville S, Cleguer C, Garrigue C (2022) Ecoregional and temporal dynamics of dugong habitat use in a complex coral reef lagoon ecosystem. Scientific Reports 12, 552.
| Crossref | Google Scholar |
Evans PGH, Hammond PS (2004) Monitoring cetaceans in European waters. Mammal Review 34, 131-156.
| Crossref | Google Scholar |
Faria DM, Steel D, Baker CS, da Silva JM, de Meirelles ACO, Souto LRA, Siciliano S, Barbosa LA, Secchi E, Di Tullio JC, de Oliveira LR, Ott PH, Farro APC (2022) Mitochondrial diversity and inter-specific phylogeny among dolphins of the genus Stenella in the Southwest Atlantic Ocean. PLoS ONE 17, e0270690.
| Crossref | Google Scholar |
Garrigue C, Derville S (2019) Maracas 7. RV Alis. Available at https://doi.org/10.17600/18000892
Garrigue C, Fernandez JM, Badie JM, Bernard C, Greaves J, Rivaton J, Trescinski M (2000) Impact of the human activities in short-finned pilot whales (Globicephala macrorhynchus) and pygmy sperm whale (Kogia breviceps) of the South West Pacific ocean by measuring Cs-137, K-40 and Pb-210. Environmental Changes and Radioactive Tracers. IRD Editions, Paris, 49-58.
Garrigue C, Greaves J, Chambellant M (2001) Characteristics of the New Caledonian Humpback whale population. Memoirs of the Queensland Museum 47, 69-75.
| Google Scholar |
Garrigue C, Clua E, Breitenstein D (2003) Identification of a juvenile pygmy blue whale (Balaenoptera musculus brevicauda) in New Caledonia, South-West Pacific. Report SC/55/SH4 to the Scientific Committee of the International Whaling Commission. 26 May–6 June 2003. Berlin, Germany. (International Whaling Commission)
Garrigue C, Patenaude N, Marsh H (2008) Distribution and abundance of the dugong in New Caledonia, southwest Pacific. Marine Mammal Science 24, 81-90.
| Crossref | Google Scholar |
Garrigue C, Oremus M, Dodémont R, Bustamante P, Kwiatek O, Libeau G, Lockyer C, Vivier JC, Dalebout ML (2016) A mass stranding of seven Longman’s beaked whales (Indopacetus pacificus) in New Caledonia, South Pacific. Marine Mammal Science 32, 884-910.
| Crossref | Google Scholar |
Garrigue C, Bonneville C, Cleguer C, Oremus M (2022) Extremely low mtDNA diversity and high genetic differentiation reveal the precarious genetic status of dugongs in New Caledonia, South Pacific. Journal of Heredity 113, 516-524.
| Crossref | Google Scholar |
Hamel M, Marsh H, Cleguer C, Garrigue C, Oremus M (2022) Dugong dugon (New Caledonia subpopulation). The IUCN Red List of Threatened Species. Version 2022. Available at www.iucnredlist.org
Hénin C, Cresswell GR (2005) Upwelling along the western barrier reef of New Caledonia. Marine and Freshwater Research 56, 1005-1010.
| Crossref | Google Scholar |
Hildebrand JA, Frasier KE, Baumann-Pickering S, Wiggins SM, Merkens KP, Garrison LP, Soldevilla MS, McDonald MA (2019) Assessing seasonality and density from passive acoustic monitoring of signals presumed to be from pygmy and dwarf sperm whales in the gulf of Mexico. Frontiers in Marine Science 6, 66.
| Crossref | Google Scholar |
Hill MC, Bendlin AR, Van Cise AM, Milette-Winfree A, Ligon AD, Adam C, Deakos MH, Oleson EM (2019) Short-finned pilot whales (Globicephala macrorhynchus) of the Mariana Archipelago: individual affiliations, movements, and spatial use. Marine Mammal Science 35, 797-824.
| Crossref | Google Scholar |
Hobson KA (1999) Tracing origins and migration of wildlife using stable isotopes: a review. Oecologia 120, 314-326.
| Crossref | Google Scholar | PubMed |
Kemper C, Gibbs P, Obendorf D, Marvanek S, Lenghaus C (1994) A review of heavy metal and organochlorine levels in marine mammals in Australia. Science of The Total Environment 154, 129-139.
| Crossref | Google Scholar | PubMed |
Kojadinovic J, Jackson CH, Cherel Y, Jackson GD, Bustamante P (2011) Multi-elemental concentrations in the tissues of the oceanic squid Todarodes filippovae from Tasmania and the southern Indian Ocean. Ecotoxicology and Environmental Safety 74, 1238-1249.
| Crossref | Google Scholar | PubMed |
Laist DW, Knowlton AR, Mead JG, Collet AS, Podesta M (2001) Collisions between ships and whales. Marine Mammal Science 17, 35-75.
| Crossref | Google Scholar |
Lanyon JM, Athousis C, Sneath HL, Burgess EA (2021) Body scarring as an indicator of social function of dugong (Dugong dugon) tusks. Marine Mammal Science 37, 962-981.
| Crossref | Google Scholar |
Laran S, Van Canneyt O, Doremus G, Garrigue C, Berr T, Bourgogne H, Genu M, Spitz J, Ridoux V (2023) Who lives in the open sea? Distribution and densities of surfacing Marine megafauna in three territories from the southwestern Pacific region (New Caledonia, Wallis-Futuna and French Polynesia). Pacific Conservation Biology
| Crossref | Google Scholar |
Lockyer C, Garrigue C (2021) Age estimation from teeth in Longman’s beaked whales (Indopacetus pacificus) stranded in New Caledonia (South Pacific). Cetacean Population Studies 3, 189-197.
| Crossref | Google Scholar |
Lyrholm T, Leimar O, Gyllensten U (1996) Low diversity and biased substitution patterns in the mitochondrial DNA control region of sperm whales: implications for estimates of time since common ancestry. Molecular Biology and Evolution 13(10), 1318-1326.
| Crossref | Google Scholar | PubMed |
Lyrholm T, Leimar O, Johanneson B, Gyllensten U (1999) Sex-biased dispersal in sperm whales: contrasting mitochondrial and nuclear genetic structure of global populations. Proceedings of the Royal Society of London, Series B: Biological Sciences 266(1417), 347-354.
| Crossref | Google Scholar |
Mahaffy SD, Baird RW, McSweeney DJ, Webster DL, Schorr GS (2015) High site fidelity, strong associations, and long-term bonds: short-finned pilot whales off the island of Hawai‘i. Marine Mammal Science 31(4), 1427-1451.
| Crossref | Google Scholar |
Manceau A, Gaillot A-C, Glatzel P, Cherel Y, Bustamante P (2021a) In vivo formation of HgSe nanoparticles and Hg-tetraselenolate complex from methylmercury in seabirds-implications for the Hg-Se antagonism. Environmental Science & Technology 55, 1515-1526.
| Crossref | Google Scholar | PubMed |
Manceau A, Azemard S, Hédouin L, Vassileva E, Lecchini D, Fauvelot C, Swarzenski PW, Glatzel P, Bustamante P, Metian M (2021b) Chemical forms of mercury in blue marlin billfish: implications for human exposure. Environmental Science & Technology Letters 8, 405-411.
| Crossref | Google Scholar |
Marsh H (2009) Dugong: Dugong dugon. In ‘Encyclopaedia of marine mammals’. 2nd edn. (Eds WF Perrin, B Würsig, JGM Thewissen) pp. 332–335. (Academic Press: London) https://doi.org/10.1016/b978-0-12-373553-9.00080-8
Marsh H, Channells PW, Heinsohn GE, Morissey J (1982) Analysis of stomach contents of dugongs from Queensland. Australian. Wildlife Research 9, 55-67.
| Crossref | Google Scholar |
Martoja R, Berry JP (1980) Identification of tiemannite as a probable product of demethylation of mercury by selenium in cetaceans. Vie et Milieu 30, 7-10 Available at https://hal.sorbonne-universite.fr/hal-03007793.
| Google Scholar |
Mcalpine DF, Murison LD, Hoberg EP (1997) New records for the pygmy sperm whale, Kogia breviceps (Physeteridae) from Atlantic Canada with notes on diet and parasites. Marine Mammal Science 13, 701-704.
| Crossref | Google Scholar |
Meland K, Aas PØ (2013) A taxonomical review of the Gnathophausia (Crustacea, Lophogastrida), with new records from the northern mid-Atlantic ridge. Zootaxa 3664, 199-225.
| Crossref | Google Scholar | PubMed |
Menkes CE, Allain V, Rodier M, Gallois F, Lebourges-Dhaussy A, Hunt BPV, Smeti H, Pagano M, Josse E, Daroux A, Lehodey P, Senina I, Kestenare E, Lorrain A, Nicol S (2015) Seasonal oceanography from physics to micronekton in the south-west Pacific. Deep Sea Research Part II: Topical Studies in Oceanography 113, 125-144.
| Crossref | Google Scholar |
Meynecke J-O, Meager JJ (2016) Understanding strandings: 25 years of Humpback Whale (Megaptera novaeangliae) strandings in Queensland, Australia. Journal of Coastal Research 75, 897-901.
| Crossref | Google Scholar |
Mitchell J (1978) Incremental growth layers in the dentine of dugong incisors (Dugong dugon (Müller)) and their application to age determination. Zoological Journal of the Linnean Society 62, 317-348.
| Crossref | Google Scholar |
Nigro M, Leonzio C (1996) Intracellular storage of mercury and selenium in different marine vertebrates. Marine Ecology Progress Series 135, 137-143.
| Crossref | Google Scholar |
Olu K, Allain V (2020) Kanarecup cruise. RV Alis. Available at https://doi.org/10.17600/18001103
Oremus M, Gales R, Kettles H, Baker CS (2013) Genetic evidence of multiple matrilines and spatial disruption of Kinship bonds in mass strandings of long-finned pilot whales, Globicephala melas. Journal of Heredity 104, 301-311.
| Crossref | Google Scholar | PubMed |
Pelletier E (1986) Mercury-selenium interactions in aquatic organisms: a review. Marine Environmental Research 18, 111-132.
| Crossref | Google Scholar |
Peltier H, Dabin W, Daniel P, Van Canneyt O, Dorémus G, Huon M, Ridoux V (2012) The significance of stranding data as indicators of cetacean populations at sea: modelling the drift of cetacean carcasses. Ecological Indicators 18, 278-290.
| Crossref | Google Scholar |
Quetglas A, de Mesa A, Ordines F, Grau A (2010) Life history of the deep-sea cephalopod family Histioteuthidae in the western Mediterranean. Deep Sea Research Part I: Oceanographic Research Papers 57, 999-1008.
| Crossref | Google Scholar |
R Core Team (2023) ‘R: a language and environment for statistical computing.’ (R Foundation for Statistical Computing: Vienna, Austria) Available at http://www.r-project.org/
Rahman IHA, Chua-Anusorn W, Webb J, Macey DJ, Pierre TGS (1999) Characterization of dugong liver ferritin. Analytica Chimica Acta 393, 235-243.
| Crossref | Google Scholar |
Rancurel P (1973) Quelques échouages de cétacés survenus dans le sud-ouest Pacifique en 1972. Bulletin du Pacifique Sud 3, 12-15.
| Google Scholar |
Robineau D, Rancurel P (1981) Sur deux spécimens du genre Kogia (Cetacea, Physeteridae) en Nouvelle-Calédonie. Zeitschrift fur Säugetiertunde 46, 56-58.
| Google Scholar |
Rodrigue A, Effantin G, Mandrand-Berthelot M-A (2005) Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. Journal of Bacteriology 187, 2912-2916.
| Crossref | Google Scholar | PubMed |
Santos MB, Pierce GJ, López A, Reid RJ, Ridoux V, Mente E (2006) Pygmy sperm whales Kogia breviceps in the Northeast Atlantic: new information on stomach contents and strandings. Marine Mammal Science 22, 600-616.
| Crossref | Google Scholar |
Stadler JA, Schweyen RJ (2002) The yeast iron regulon is induced upon cobalt stress and crucial for cobalt tolerance. Journal of Biological Chemistry 277, 39649-39654.
| Crossref | Google Scholar | PubMed |
Staudinger MD, McAlarney RJ, McLellan WA, Ann Pabst D (2014) Foraging ecology and niche overlap in pygmy (Kogia breviceps) and dwarf (Kogia sima) sperm whales from waters of the U.S. mid-Atlantic coast. Marine Mammal Science 30, 626-655.
| Crossref | Google Scholar |
Sylvestre JP (1988) On a specimen of pygmy sperm whale Kogia breviceps (Blainville, 1838) from New Caledonia. Aquatic Mammals 14(2), 76-77.
| Google Scholar |
Unger B, Rebolledo ELB, Deaville R, Gröne A, IJsseldijk LL, Leopold MF, Siebert U, Spitz J, Wohlsein P, Herr H (2016) Large amounts of marine debris found in sperm whales stranded along the North Sea coast in early 2016. Marine Pollution Bulletin 112, 134-141.
| Crossref | Google Scholar | PubMed |
Van Canneyt O, Dorémus G, Garrigue C, Hébert O, Laran S, Watremez P, Ridoux V (2016) New insight on marine mammals of the Southwest Pacific ocean: Part I- Large aerial survey (REMMOA) in the New Caledonia EEZ. Poster session presented at 30th Annual conference of the European Cetacean Society. (Funchal, Portugal)
Van Cise AM, Baird RW, Baker CS, Cerchio S, Claridge D, Fielding R, Hancock-Hanser B, Marrero J, Martien KK, Mignucci-Giannoni AA, Oleson EG, Oremus M, Poole MM, Rosel PE, Taylor BL, Morin PA (2019) Oceanographic barriers, divergence, and admixture: phylogeography and taxonomy of two putative subspecies of short - finned pilot whale. Molecular Ecology 28(11), 2886-2902.
| Crossref | Google Scholar | PubMed |
Virgili A, Authier M, Boisseau O, Cañadas A, Claridge D, Cole T, Corkeron P, Dorémus G, David L, Di-Méglio N, Dunn C, Dunn TE, García-Barón I, Laran S, Lauriano G, Lewis M, Louzao M, Mannocci L, Martínez-Cedeira J, Palka D, Panigada S, Pettex E, Roberts JJ, Ruiz L, Saavedra C, Santos MB, Van Canneyt O, Vázquez Bonales JA, Monestiez P, Ridoux V (2018) Combining multiple visual surveys to model the habitat of deep - diving cetaceans at the basin scale. Global Ecology and Biogeography 28, 300-314.
| Crossref | Google Scholar |
Wahoulo E (2019) Développement d’une application web de signalisation des échouages de mammifères marins en Nouvelle-Calédonie (1.0). Rapport de stage de Licence Informatique, Université de la Nouvelle-Calédonie. p. 36. (Université de la Nouvelle-Calédonie: Zenodo) Available at https://doi.org/10.5281/zenodo.7758538
West KL, Walker WA, Baird RW, White W, Levine G, Brown E, Schofield D (2009) Diet of pygmy sperm whales (Kogia breviceps) in the Hawaiian Archipelago. Marine Mammal Science 25, 931-943.
| Crossref | Google Scholar |
Whitehead H (2018) Sperm whale: Physeter macrocephalus. In ‘Encyclopedia of marine mammals’. (Eds W Perrin, B Würsig, J Thewissen) pp. 919–925. (Academic Press) https://doi.org/10.1016/b978-0-12-804327-1.00242-9
Whitehead H, Dillon M, Dufault S, Weilgart L, Wright J (1998) Non-geographically based population structure of South Pacific sperm whales: dialects, fluke-markings and genetics. Journal of Animal Ecology 67, 253-262.
| Crossref | Google Scholar |
Young RE, Vecchione M (2009) Ornithoteuthis Okada 1927. Bird squid. Version 29 November 2009 (under construction). Available at http://tolweb.org/Ornithoteuthis/19942/2009.11.29 in The Tree of Life Web Project, http://tolweb.org/


