DNA barcoding reveals larval fish diversity and distribution along the Cibareno River (West Java, Indonesia)
Arif Wibowo A , Andi Chadijah A , Kurniawan Kurniawan A B C , Vitas Atmadi Prakoso A B C * , Dwi Atminarso
A B C * , Dwi Atminarso  A B C , Deni Irawan A , Fathur Rochman
A B C , Deni Irawan A , Fathur Rochman  A , Septiana Sri Astuti A , Indah Lestari Surbani D , Tri Deniansen A , Imron Rosadi E , Yohanes Yudha P. Jaya E , Sudarsono Sudarsono E , Lee J. Baumgartner B , Nicolas Hubert
A , Septiana Sri Astuti A , Indah Lestari Surbani D , Tri Deniansen A , Imron Rosadi E , Yohanes Yudha P. Jaya E , Sudarsono Sudarsono E , Lee J. Baumgartner B , Nicolas Hubert  F and Ivor G. Stuart B
F and Ivor G. Stuart B
A
B
C
D
E
F
Abstract
There is a global problem with ongoing riverine infrastructure projects where, despite knowledge of potential environmental impacts, there is rapid development, often without appropriate environmental safeguards. This results in fragmentation of riverine fish communities, especially diadromous species. Understanding freshwater fish larval ecology is critical to provide insight into the likely impacts of these projects.
To assess fish larval biodiversity on the basis of DNA barcoding, abundance and its distribution pattern in the Cibareno River.
Fish larvae were collected at six locations in the Cibareno River. The larvae were identified by DNA barcoding.
A notable disparity was seen in the distribution of larval abundance in different locations. The non-native species, Poecilia reticulata, was the most abundant larval species, with an intraspecific diversity of 0.003 (99.7% similarity). The upstream area exhibited a lower level of larval species diversity than did the downstream area.
Genetic identification can reliably identify fish larvae and determine their spatial riverside distribution in the Cibareno River. The conservation of connectivity maintains fish community integrity and diversity between upstream and downstream locations in the weir building plan.
This discovery emphasises the relevance of larval identification in fish biodiversity assessment and sustainable fisheries resource monitoring.
Keywords: aquatic ecology, biodiversity, conservation, DNA barcoding, early life stages, ichthyoplankton, larval fish, species composition.
Introduction
River flow regulation is a major threat to riverine fish communities, with population declines occurring globally (Darwall and Freyhof 2015). Among the chief impacts of new infrastructure is an immediate change to upstream hydrodynamics from dynamic free-flowing to deep slow-flowing habitats, with a consequent loss of fish biodiversity (Ward and Stanford 1995; Widén et al. 2021). Fragmentation of the riverine continuum also has impacts on upstream and downstream fish migrations, with diadromous fish communities typically declining above barriers with varying degrees of isolation on freshwater fishes in upstream area (Drouineau et al. 2018). When the river flow is dammed, the resultant conditions can be more favourable to introduced species than native species (Mercado-Silva et al. 2009).
South-east Asia is emblematic of the global trend for ongoing riverine infrastructure projects where, despite strong existing knowledge of potential serious environmental impacts, especially to riverine fish communities, there is rapid development, often without appropriate environmental safeguards (King and Brown 2010; Soukhaphon et al. 2021). In some cases, fisheries resources are in serious decline (Campbell and Barlow 2020; Vu et al. 2021), highlighting the need for much greater scientific effort to quantify these changes so as to target efforts to protect key elements of riverine hydrology and fish communities (Schmutz et al. 2015; Ziegeweid et al. 2022).
Early life stages (e.g. eggs and larvae) of riverine fish are the most vulnerable to perturbation, while also playing a crucial trophic link between plankton and higher predators, as well as in recruitment and self-sustaining populations (Heimeier et al. 2010; Ardura et al. 2016). A comprehensive understanding of the biology of early fish life-history stages is necessary to improve fisheries management (Kouhanestani et al. 2020), which can help protect critical habitats and species (Lucas and Baras 2000; Cooke 2008). Specifically, knowledge of larval ecology can provide fundamental information on fish reproductive biology, such as reproduction timing, geo-locations, migration routes, and population recruitment success (Cooke et al. 2016; Wibowo et al. 2017). Such information is very important and extremely valuable for understanding the impacts of riverine infrastructure, developing management and conservation plans, establishing fishing management strategies, and contributing to the conservation of endangered and vulnerable species (Reynalte-Tataje et al. 2011; Wibowo et al. 2017).
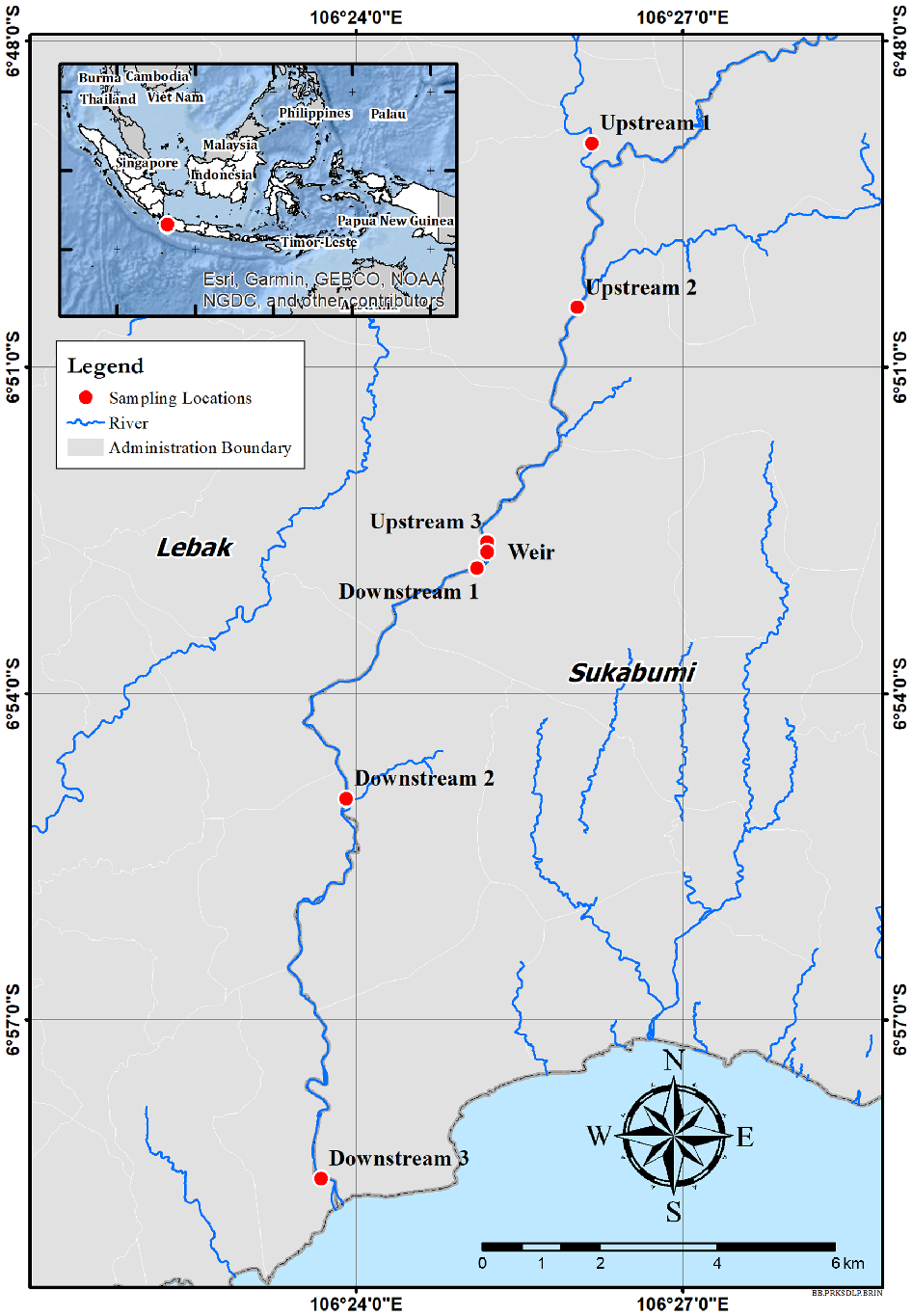
The Cibareno River in West Java, Indonesia forms a natural border between Banten and West Java provinces in Indonesia. The upstream area is located in the Halimun National Park area of West Java. Cibareno River flows ~37 km to Pelabuhan Ratu Bay. There is no recent information available to evaluate the fish biodiversity in the river. In 2002, ~22 fish species from 10 families were recorded (Rachmatika et al. 2002). Recent assessments with DNA barcoding of the ichthyofauna of Java Island reported 159 species (Dahruddin et al. 2017; Hubert et al. 2019), suggesting that fish diversity in the Cibareno River might be underestimated. An irrigation weir project is currently under development in the middle part of the Cibareno River (Fig. 1). While conducting the survey, the dam foundation had reached a height of ~1 m, a fraction of the planned 3.25 m, stretching across the entire riverbed from left to right. This may constitute a barrier for upstream migration of fish early stages, a situation of concern for the Halimun National Park fish communities. The objective of the present study was to characterise at the species level the composition of the ichthyoplankton of the Cibareno River at various elevations and its variation during the course of 1 year. The identification of fish larvae is challenging if based on morphological characters, because of a general lack of diagnostic characters (Teletchea 2009; Ko et al. 2013). However, DNA barcoding, the use of the mitochondrial cytochrome oxidase I (COI) gene as a species tag for molecular identification, has been successfully used to identify fish larvae to the species level and characterise ichthyoplankton (Hubert et al. 2010, 2015; Wibowo et al. 2017; Hulley et al. 2018). Molecular techniques offer unprecedented insights on identification of fish larvae in aquatic environments (Chu et al. 2019; Lira et al. 2023).
Map of the study location in the Cibareno River, Sukabumi, West Java, Indonesia, with three fish sampling locations upstream and downstream of Caringin Weir. Upstream 1, 6.8157°S, 106.4361°E; Upstream 2, 6.8409°S, 106.4338°E; Upstream 3, 6.8768°S, 106.4200°E; Weir, 6.8783°S, 106.4200°E; Downstream 1, 6.8808°S, 106.4185°E; Downstream 2, 6.9161°S, 106.3985°E; and Downstream 3, 6.9743°S, 106.3946°E.
The present study makes use of the previously published DNA barcode reference library of the freshwater fishes of Java (Dahruddin et al. 2017) to document the species composition of the ichthyoplankton occurring in the Cibareno River over 1 year, by using DNA barcoding. In this study, DNA barcoding was essential for accurately identifying fish larval species in the Cibareno River because traditional methods based on morphological characteristics can be challenging. We used standardised fishing methods to examine larval community composition along the Cibareno River.
Materials and methods
Study sites
The 37-km-long Cibareno River originates in the Gunung Halimun National Park and flows south between Banten and West Java provinces through a forested and conserved catchment to the Indian Ocean, west of the Pelabuhan Ratu Bay (Rachmatika et al. 2002). The steep-gradient catchment is characterised by rocky substrates, waterfalls and swift water. Caringin Weir, in Sukabumi District, is a single weir that may disrupt fish movement, including that of high-value fish such as Tor tambra (Amanda et al. 2023). During this study, the weir was under construction and its foundation was elevated 1 m (the final height will be 3.25 m) above the riverbed, extending from one bank to the other.
Specimen collection
Larval and adult fish specimens were collected at six different locations along the Cibareno River, three of which were upstream of the location of the Caringin weir project and the other three were downstream (Fig. 1). Upstream and downstream sampling locations were 0, 5 and 10 km from the location of the weir.
The experiment was conducted under animal research and ethic approval for ‘Fish Biodiversity and Environmental Assessment of Cibareno River for Supporting Inland Fisheries and Fish Domestication’ number 60.1/BRSDM-BRPBATPP/LB.200/I/2022 granted by the Research Institute of Freshwater Aquaculture and Fisheries Extension, Ministry of Marine Affairs and Fisheries, Republic of Indonesia.
The following four types of fishing gear were consistently used to collect adult fish for each sampling event: (i) five multi-panel monofilament gillnets (each 5 m wide, with mesh stretched width of 19, 25, 37, 50 and 75 mm), (ii) ten collapsible bait traps (400 × 220 × 220 mm), (iii) ten cast nets (2-m diameter, 25-mm mesh) and (iv) two single wing fyke nets (wing, pairs of 4 m long, 0.2 m in diameter, net body 1.25 m long, 7-mm stretched mesh). All fish caught were photographed, measured (to the nearest millimetre) and weighed (to the nearest 0.1 g). All experimental fishing was conducted between 07:00 and 14:00 hours during 5 days. To capture any potential seasonal variations in fish species and relative abundance, all six sites were sampled three times during the wet season and three times during the dry season (between March and September 2022). Individuals were further identified to the species level by using monographs and field guides (Kottelat et al. 1993; Rachmatika et al. 2002; Keith et al. 2015), and further confirmed by a visual examination of specimen photographs of the Java fish species catalogued in BOLD (Dahruddin et al. 2017; Keith et al. 2017; Hubert et al. 2019).
At the same six sites, preflexion (i.e. from hatching to the onset of notochord flexion) to postflexion (i.e. from formation of the caudal fin to attainment of complete development fin rays and scales) larval fish (Leis and Carson-Ewart 2000) were sampled with a 30-cm diameter modified bongo net (0.3-mm mesh). The net was kept submerged ~5 cm below the surface for two sessions of 15 min, so as to collect replicates at each site. These sampling sessions were conducted early in the morning (from 07:00 to 08:00 hours). Larvae were kept in water after collection, and manually sorted. To reduce animal suffering, fish larvae were euthanised with anesthetic overdose (Eugenol; dose: 4 mL per 1 L of water) at the collection site. The procedure for larval storage and sorting followed Wibowo et al. (2017) with slight modifications. Each fish larva was stored in a 1.5-mL tube containing absolute ethanol. Each tube was labelled and prepared, and anesthetised larvae were photographed alongside the tubes before storing in the tubes. Larval counts were conducted by sorting similar larvae and grouping them together, facilitating the species identification and tallying the number of larvae at each station. Larvae were selected for DNA analysis from each group suspected to belong to distinct species and species composition among larval fish community was examined using multi-dimensional scaling (MDS) analysis with the vegan package (ver. 2.6-8, J. Oksanen et al., see https://cran.r-project.org/package=vegan/) in RStudio (ver. 1.2.1335, Posit Software, PBC, Boston, MA, USA, see https://posit.co/products/open-source/rstudio/). This analysis was applied to log-transformed abundance estimates to visualise variations in the composition occurring through space (sampling locations) and time (seasons).
Extraction, amplification and sequencing of DNA
Total genomic DNA was isolated using a genomic DNA extraction kit (Geneaid Tissue Genomic DNA Mini Kit GT050, Taiwan) from either muscle tissue (for adult fish) or entire larvae, following the protocol provided by the manufacturer (Wibowo et al. 2021). A 652 bp fragment of the mitochondrial cytochrome c oxidase subunit-1 gene (COI) was amplified using the primers Fish-COI-F (5′-TAA TAC GAC TCA CTA TAG GGT TCT CCA CCA ACC ACA ARG AYA TYGG-3) and COI-Fish-R (5′-ATT AAC CCT CAC TAA AGG GCA CCT CAG GGT GTC CGA ARA AYC ARAA-3′) (Ivanova et al. 2007).
The COI fragment was amplified in a 12.5-μL reaction volume that included 4.0 μL of ultrapure water, 0.625 μL of each primer (1 mM), 6.25 μL of 2× QIAGEN Multiplex PCR Master Mix (QIAGEN, Germany), and 1 μL of DNA template (~100 ng L−1). Polymerase chain reaction (PCR) cycling included an initial 15-min DNA polymerase denaturation stage at 95°C, 35 cycles of 30 s at 94°C, 90 s at 55°C, and 30 s at 72°C, and a 5-min extension at 72°C. PCR products were detected on a 1% agarose gel and purified with the ASAP PCR clean-up kit (ArcticZymes). The EZ-Seq service (Macrogen, Singapore) performed the sequencing and all sequences were submitted to GenBank (PP352528–PP 352565).
DNA barcoding and specimen identification
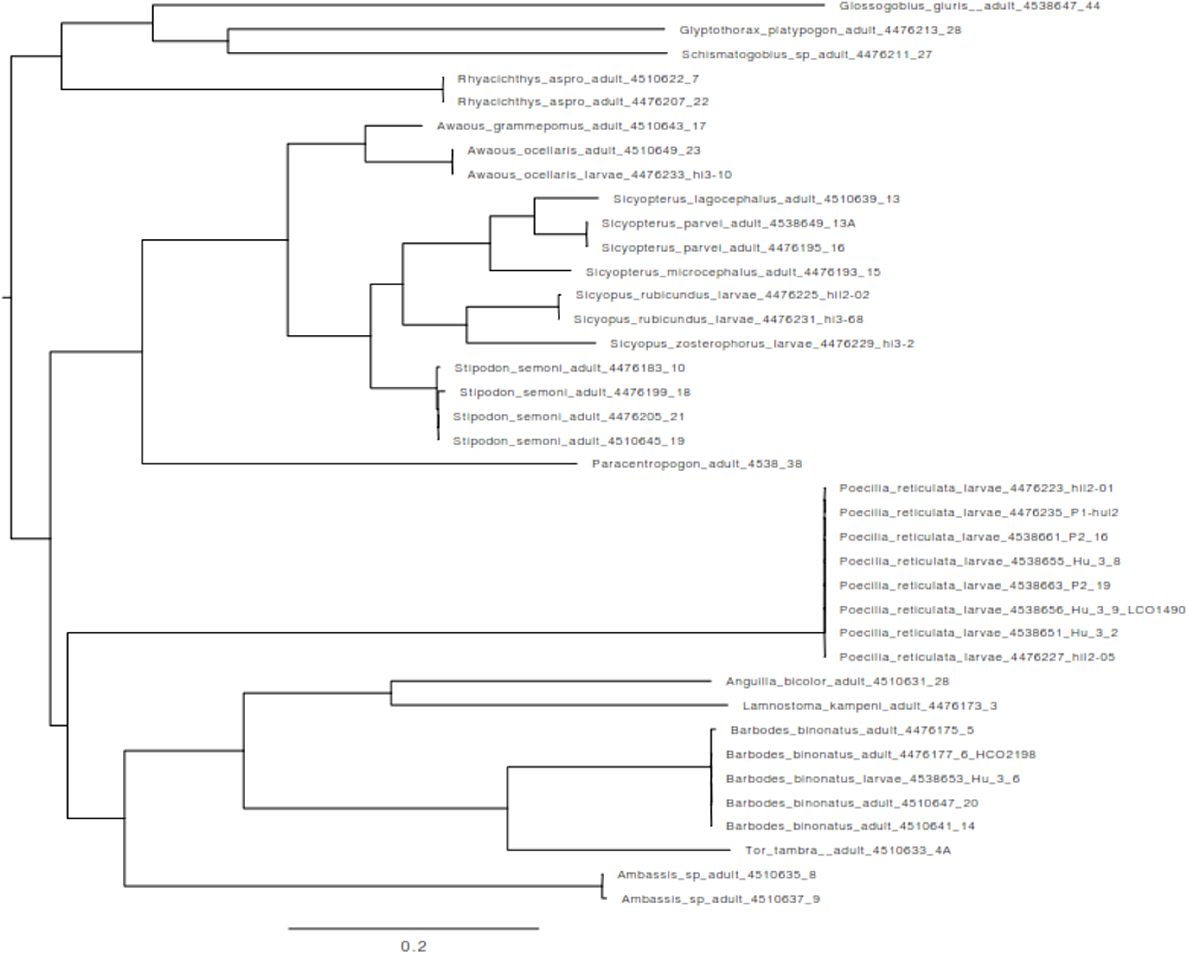
Chromatograms were inspected manually and trimmed to 652 bp, and a multiple-sequence alignment was performed using Geneious Prime (ver. 2024.0.1, see https://www.geneious.com; Kearse et al. 2012). Individual sequences were identified to the species level through basic local alignment search tool (BLAST) searches in NCBI GenBank database (see http://www.nlm.nih.gov/). Sequences were assigned to a known taxon when a match above 98% similarity was found (Hebert et al. 2003; Ross et al. 2008). Additional sequences available in GenBank for the taxa identified were downloaded and jointly analysed with the newly generated sequences. The final multiple sequence alignment was subsequently used to construct a phylogenetic tree for a visual inspection of the genetic divergence among sequences. We reconstructed a Maximum Likelihood (ML) tree using IQ-TREE (ver. 1.6.12, see http://iqtree.cibiv.univie.ac.at; Nguyen et al. 2015; Trifinopoulos et al. 2016) with the most-likely substitution model according to ModelFinder (see http://www.iqtree.org/ModelFinder/; Kalyaanamoorthy et al. 2017) following the Bayesian information criterion (BIC).
Results
Fish identification
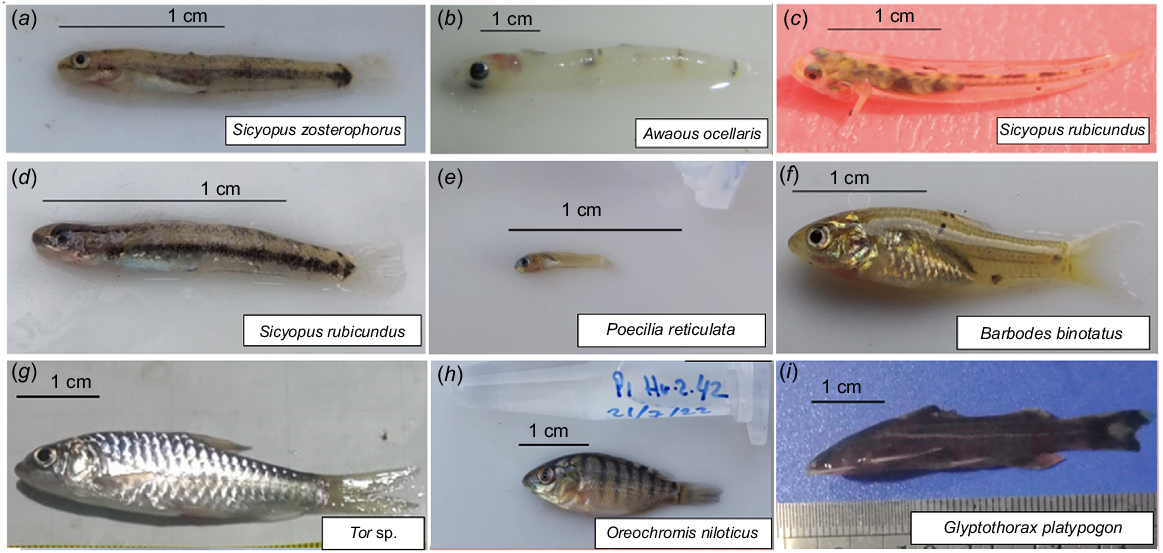
In total, 800 fish (including larvae and juveniles) were collected from six locations over the 6-month period encompassing the wet and dry seasons. Among these 800 fish, 9 morphotypes were detected (Fig. 2).
Photographs of specimens representing morphotypes detected in the (a–f) fish larvae and (g–i) juveniles collected in the Cibareno River, West Java, Indonesia.
In total, 80 chromatogram (40 forward and reverse pairs) sequences were obtained from 40 individuals, including adult specimens and representative samples of the larval morphotypes identified. We did not perform molecular testing on the Oreochromis niloticus larval samples because our focus was on native fish species that had not yet been identified. In addition, the O. niloticus larvae can be clearly identified on the basis of their morphological characteristics. Sequencing failed for only 2 individuals and 38 sequences, in total, were produced, with a sequence length of 628–632 base pairs, with 1 to 8 specimens per morphotype. These 38 sequences were subsequently identified to the species level by using the BLAST procedure in GenBank (Table 1). All sequences were unambiguously identified to the species level with the 98% similarity threshold, excepting Ambassis sp., Schismatogobius sp. and Paracentropogon sp.
| Number | Status | Best match | Similarity (%) | Nearest neighbour | Similarity (%) | |
|---|---|---|---|---|---|---|
| 1 | Adult | Ambassis sp. | 99.84 | Ambassis gymnocephalus | 92.57 | |
| 2 | Adult | Ambassis sp. | 99.68 | Ambassis gymnocephalus | 92.40 | |
| 3 | Adult | Anguilla bicolor | 100 | Anguilla obscura | 96.02 | |
| 4 | Adult | Awaous grammepomus | 99.83 | Awaous ocellaris | 99.84 | |
| 5 | Adult | Awaous ocellaris | 99.82 | Awaous grammepomus | 93.16 | |
| 6 | Larvae | Awaous ocellaris | 99.82 | Awaous grammepomus | 93.72 | |
| 7 | Adult | Barbodes binotatus | 99.84 | Barbodes banksi | 99.35 | |
| 8 | Adult | Barbodes binotatus | 100 | Barbodes banksi | 99.35 | |
| 9 | Adult | Barbodes binotatus | 100 | Barbodes banksi | 99.35 | |
| 10 | Adult | Barbodes binotatus | 100 | Barbodes banksi | 99.35 | |
| 11 | Larvae | Barbodes binotatus | 100 | Barbodes banksi | 99.35 | |
| 12 | Adult | Glossogobius giuris | 99.67 | Awaous melanocephalus | 92.94 | |
| 13 | Larvae | Glyptothorax platypogon | 100 | Glyptothorax robustus | 98.53 | |
| 14 | Adult | Lamnostoma kampeni | 100 | Lamnostoma mindora | 97.07 | |
| 15 | Adult | Paracentropogon | NA | Inimicus japonicus | 87.26 | |
| 16 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 17 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 18 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 19 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 20 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 21 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 22 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 23 | Larvae | Poecilia reticulata | 100 | Poecilia wingei | 100 | |
| 24 | Adult | Rhyacichthys aspro | 100 | Rhyacichthys gilberti | 95.22 | |
| 25 | Adult | Rhyacichthys aspro | 100 | Rhyacichthys gilberti | 95.22 | |
| 26 | Adult | Schismatogobius sp. A | NA | Schismatogobius insignus | 93.28 | |
| 27 | Adult | Sicyopterus lagocephalus | 99.02 | Sicyopterus marquesensis | 96.18 | |
| 28 | Adult | Sicyopterus microcephalus | 100 | Sicyopterus pugnans | 95.06 | |
| 29 | Adult | Sicyopterus parvei | 99.35 | Sicyopterus cynocephalus | 95.32 | |
| 30 | Adult | Sicyopterus parvei | 99.35 | Sicyopterus cynocephalus | 95.32 | |
| 31 | Larvae | Sicyopus rubicundus | 100 | Sicyopus discordipinnis | 94.67 | |
| 32 | Larvae | Sicyopus rubicundus | 100 | Sicyopus discordipinnis | 94.67 | |
| 33 | Larvae | Sicyopus zosterophorus | 100 | NA | NA | |
| 34 | Adult | Stiphodon semoni | 99.68 | Stiphodon atropurpureus | 96.24 | |
| 35 | Adult | Stiphodon semoni | 99.51 | Stiphodon atropurpureus | 96.24 | |
| 36 | Adult | Stiphodon semoni | 100 | Stiphodon atropurpureus | 96.57 | |
| 37 | Adult | Stiphodon semoni | 100 | Stiphodon atropurpureus | 96.57 | |
| 38 | Larvae | Tor tambra | 99.84 | Tor tambroides | 99.68 |
BLAST analyses resulted in the detection of 19 species (Table 1), of which 8 were detected among sampled larvae, indicating that several morphotypes identified among larvae correspond to different stages of the same species (Fig. 2). These correspond to Awaous ocellaris, Sicyopus zosterophorus, Sicyopus rubicundus, Poecilia reticulata, Glyptothorax platypogon, Tor sp. and Barbodes binotatus. The ML tree reconstructed with the TIM2 + F + I + Γ model, the most likely model according to modeltest, indicated that species represented by multiple sequences represent clusters of closely related sequences (Fig. 3). At the larval stage, morphology varies widely in some species (e.g. Poecilia reticulata), making species classification difficult by using morphological characters only. The greatest number of larvae found was for P. reticulata (eight individuals).
Maximum Likelihood (ML) tree of the 38 newly generated sequences from this study, corresponding to 19 species. Sequences were generated either with adult or larval specimens.
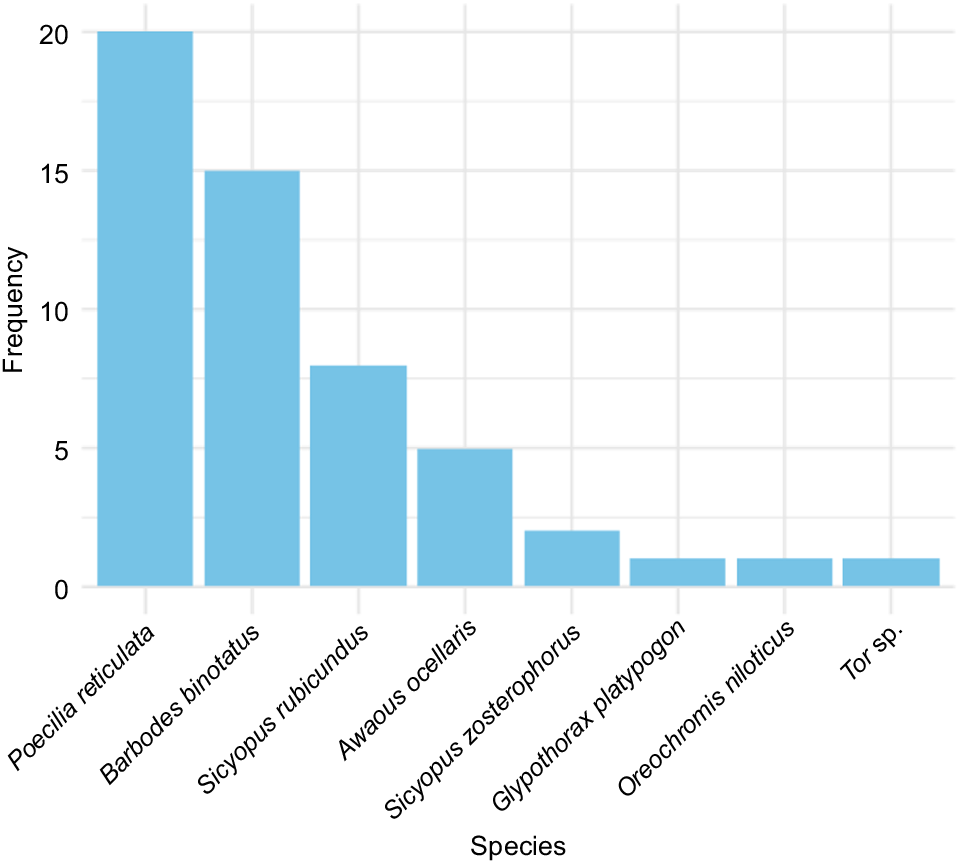
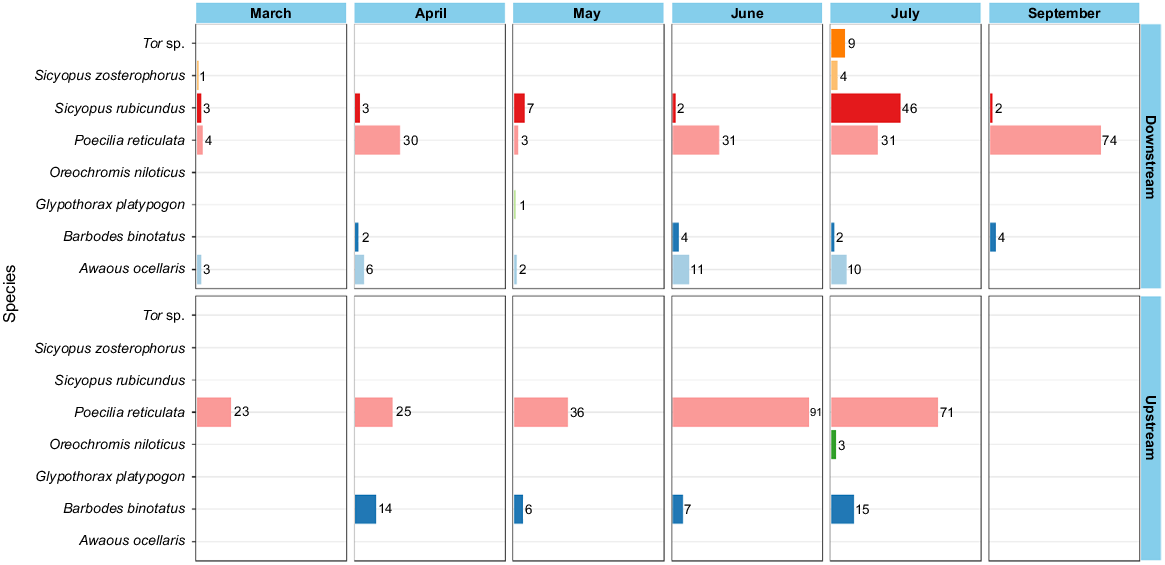
During the sampling period, Poecilia reticulata, Barbodes binotatus and Sicyopus rubicundus are the three species that were the most commonly captured in the sampling sites (Fig. 4).
Larvae of Poecilia reticulata dominated all larval assemblages because they were captured at a higher frequency than were the other species in both downstream and upstream locations (Fig. 5). Two more fish species, namely, Sicyopus rubicundus and Awaous ocellaris, were consistently observed in downstream areas during each sampling period.
Species composition of larvae sampled from the Cibareno River, West Java, Indonesia, by site and month.
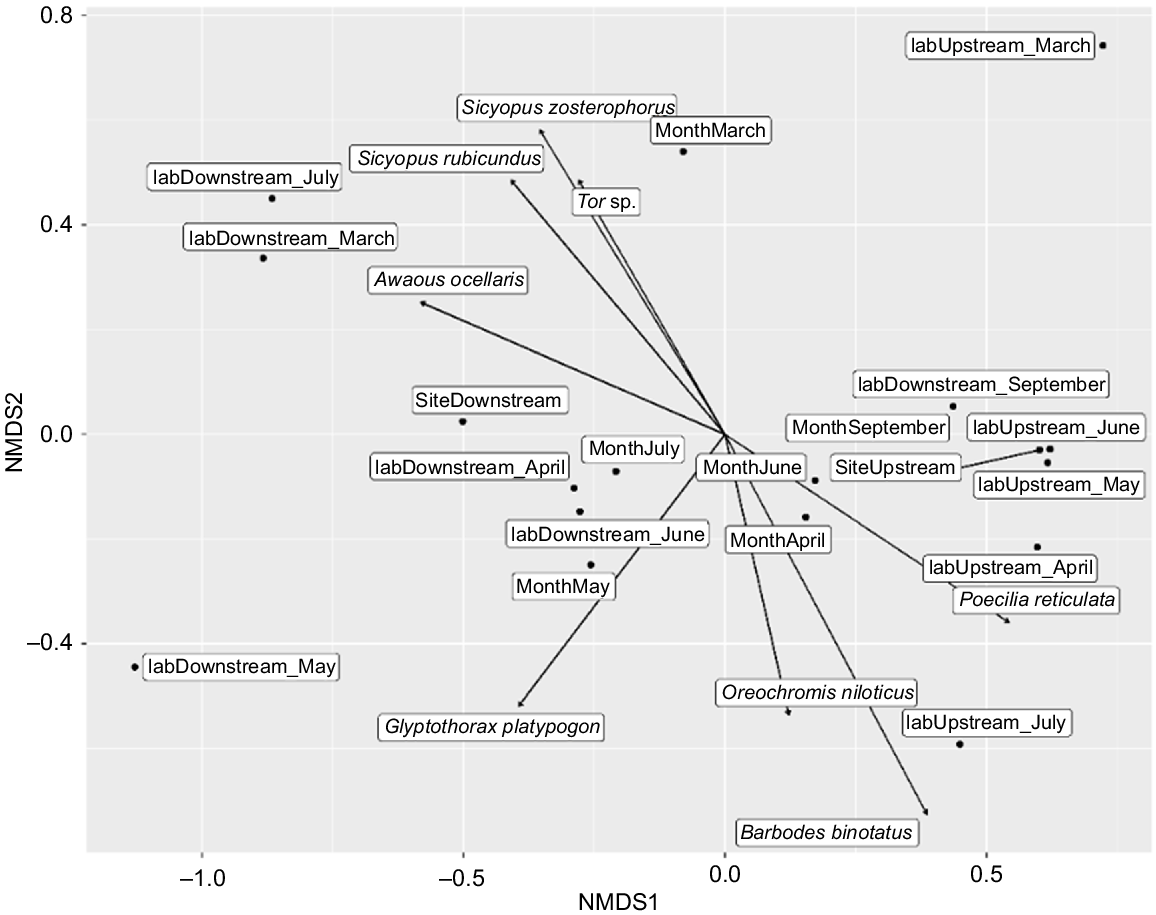
The NMDS graph shows two clusters separating upstream and downstream locations with a value of P = 0.016 (Fig. 6). The presence of larvae of Poecilia reticulata characterised the upstream sites in April and May. Barbodes binotatus larvae were characteristic of the upstream sites in July. The presence of Tor sp. and Sicyopus zosterophorus larvae in July, Glypothorax platypogon larvae in May and Awaous ocellaris larvae in March characterised the downstream area.
Discussion
Construction of riverine infrastructure is increasing throughout Indonesia, which is typical of South-east Asia, with the immediate consequence of isolating migratory fish communities (Haryani 2021; Atminarso et al. 2024). Our case study on the Cibareno River also highlighted a strong potential for restriction of diadromous fish to the river reaches downstream of Caringin Weir and upstream isolation of a potamodromous and riverine fish community.
To understand how Cibareno River fish communities responded to construction of the Caringin Weir, we first needed to validate a technique to identify fish larvae. DNA barcoding enabled us to reliably identify eight species of fish larvae in the Cibareno River, two of which (Sicyopus rubicundus and Barbodes binotatus) had not previously been reported upstream (Rachmatika et al. 2002). Additionally, we also detected the genus Tor among the larvae sampled, the species of which are high-value species only recently documented in the Cibareno catchment (Amanda et al. 2023). For rivers where there are few historical survey data, DNA barcoding is emerging as a sound technique for characterising biodiversity and for providing a baseline library for future monitoring (Hubert et al. 2015; Wibowo et al. 2017).
The central position of the Caringin Weir in the Cibareno watershed is likely to be influential for interpreting patterns of larval diversity and abundance observed among upstream and downstream sites. Elsewhere, following weir construction, there is invariably a gross simplification of upstream free-flowing lotic habitats that become deeper, slow-flowing lentic habitats (Mueller et al. 2011; Jo et al. 2019). In these modified habitats, there are usually detrimental changes to food-webs, including phytoplankton, zooplankton and ultimately native fishes (Park et al. 2023). For the Cibareno River, evaluating a broad suite of environmental variables was beyond the scope of our study but certainly we observed a greater larval species diversity downstream of Caringin Weir where free-flowing habitat remained. This river stretch was characterised by features such as pools, aquatic vegetation, woody habitat, as well as riffle and run zones, where such a broad range of habitats can support a range of fish species (Guo et al. 2017).
In tropical rivers that experience wet and dry seasons, temporal patterns of abundance in fish larvae are usually observed (Castro et al. 2005). However, within the Cibareno River, we failed to observe variation in the larval fish assemblages between March and September, which encompassed both the dry and wet seasons. The wet season is often a time of fish reproduction, when floodplain habitats become accessible and there is greater food availability for fish larvae (i.e. the flood-pulse concept) (Junk et al. 1989). Changes in species diversity and abundance of fish larvae are therefore often related to taking advantage of flow events to access areas of improved water quality, food availability, spawning habitats and for reducing predation (Chhuoy et al. 2022). However, the Cibareno River is a high-gradient system with few adjacent floodplains; hence, the more general spawning strategies of local species appears to involve adaptions to local conditions (Godfrey et al. 2017).
In efforts to biologically control malaria carrying mosquitoes, Trinidadian guppies (P. reticulata) have been introduced to many aquatic habitats in South-east Asia (Yadav and Haq 2017). Where invasive guppies become established in new aquatic habitats with few resident species, they usually induce important perturbations of local biodiversity because of their predatory nature (Morgan et al. 2004; El-Sabaawi et al. 2016; R. Froese and D. Pauly, FishBase, ver. 04/2019, see www.fishbase.org). For the Cibareno River, habitat fragmentation and hydrological habitat simplification following construction of Caringin Weir would also appear as another trigger for exacerbating guppy abundance and impacts on resident species (Alexander et al. 2015; Suryaningsih et al. 2020). In other parts of Indonesia, guppies have outcompeted local species because of faster sexual maturity, reproduction rates and population growth (Reznick et al. 2001; Herder et al. 2022).
In conclusion, we have provided a baseline fish assemblage, from which the impacts of Caringin Weir can be monitored using DNA barcoding for future monitoring, along with other field techniques. We anticipate changes in fish communities at upstream and downstream sites, which may come to light as the full impact of the new weir manifests, despite the presence of a fishway, because upstream habitats will change from free-flowing fast-water to ponded deep-water habitats. In steep, fast-flowing catchments, the water dynamics create a constant connection along the river, making larval fish communities especially sensitive to changes in their environment. We aim for our baseline data to guide infrastructure projects in South-east Asia and support the conservation of tropical river fish populations.
Data availability
Data used in this paper are organised by the first author. Access to these data can be negotiated with him.
Declaration of funding
The project funding of the study was supported by the Food and Agriculture Organization and Global Environment Facility project ‘Inland Fisheries Practices in Freshwater Ecosystems of High Conservation Value (IFish)’ (FAO project code: GCP/INS/303/GFF, GEF ID: 5759, to FAO Indonesia with salary funding for Imron Rosadi, Yohanes Yudha P. Jaya and Sudarsono Sudarsono) and the Australian Centre for International Agricultural Research (project number FIS/2018/153, with salary and travel support for Lee Baumgartner and Arif Wibowo).
Acknowledgement
The authors are grateful to the FAO Indonesia (IFish Project Team) and the Australian Centre for International Agricultural Research (ACIAR) teams for this wonderful opportunity and collaboration. The authors gratefully acknowledge Research Center for Fisheries, the Ministry of Marine Affairs and Fisheries, Republic of Indonesia, the Government Office of Fisheries, Government office of Water Resource Management, West Java, and UPTD Water Resource Management of Sukabumi, West Java. We also extend our gratitude to Boby Bagja Pratama for refining the map of our study. Finally, we thank Fery, Daman, and undergraduate students of Aquatic Resource Management, IPB University, who significantly contributed to the fieldwork.
References
Alexander ME, Kaiser H, Weyl OLF, Dick JTA (2015) Habitat simplification increases the impact of a freshwater invasive fish. Environmental Biology of Fishes 98(2), 477-486.
| Crossref | Google Scholar |
Amanda L, Prakoso VA, Boer M, Muhammad Wildan D, Kurniawan K, Rochman F, Iswantari A, Irawan D, Yudha P. Jaya Y, Rosadi I, Sudarsono S, Wibowo A (2023) Food and feeding habits of Tor tambra (Valenciennes, 1842) in Cibareno River, West Java, Indonesia. In ‘International Seminar on Fish and Fisheries Sciences (ISFFS 2023). E3S Web of Conferences, Vol. 442’, 6–7 June 2023, Denpasar, Bali, Indonesia and held virtually. (Eds CPH Simanjuntak, AR As-Syakur, Krismono, Djumanto, AM Lusiastuti, MS Baskoro, Nurjanah, HW Palm, HW Kim) Paper number 01015. (EDP Sciences) 10.1051/e3sconf/202344201015
Ardura A, Morote E, Kochzius M, Garcia-Vazquez E (2016) Diversity of planktonic fish larvae along a latitudinal gradient in the eastern Atlantic Ocean estimated through DNA barcodes. PeerJ 4, e2438.
| Crossref | Google Scholar |
Atminarso D, Baumgartner LJ, Watts RJ, Rourke ML, Bond J, Wibowo A (2024) Evidence of fish community fragmentation in a tropical river upstream and downstream of a dam, despite the presence of a fishway. Pacific Conservation Biology 30, PC22035.
| Crossref | Google Scholar |
Campbell I, Barlow C (2020) Hydropower development and the loss of fisheries in the Mekong River Basin. Frontiers in Environmental Science 8, 566509.
| Crossref | Google Scholar |
Castro MS, Bonecker ACT, Valentin JL (2005) Seasonal variation in fish larvae at the entrance of Guanabara Bay, Brazil. Brazilian Archives of Biology and Technology 48, 121-128 [In English with abstract and keywords in English and Portuguese].
| Crossref | Google Scholar |
Chhuoy S, Hogan ZS, Chandra S, Chheng P, Touch B, Utsugi K, Ngor PB (2022) Daily otolith ring validation, age composition, and origin of the endangered striped catfish in the Mekong. Global Ecology and Conservation 33, e01953.
| Crossref | Google Scholar |
Chu C, Loh KH, Ng CC, Ooi AL, Konishi Y, Huang S-P, Chong VC (2019) Utilization of DNA barcodes for the identification of larval fishes in tropical estuarine waters (Malacca Straits, Malaysia). Zoological Studies 58, 30.
| Crossref | Google Scholar |
Cooke SJ (2008) Biotelemetry and biologging in endangered species research and animal conservation: relevance to regional, national, and IUCN Red List threat assessments. Endangered Species Research 4, 165-185.
| Crossref | Google Scholar |
Cooke SJ, Martins EG, Struthers DP, Gutowsky LFG, Power M, Doka SE, Dettmers JM, Crook DA, Lucas MC, Holbrook CM, Krueger CC (2016) A moving target – incorporating knowledge of the spatial ecology of fish into the assessment and management of freshwater fish populations. Environmental Monitoring and Assessment 188(4), 239.
| Crossref | Google Scholar |
Dahruddin H, Hutama A, Busson F, Sauri S, Hanner R, Keith P, Hadiaty R, Hubert N (2017) Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species. Molecular Ecology Resources 17(2), 288-299.
| Crossref | Google Scholar | PubMed |
Darwall WRT, Freyhof J (2015) Lost fishes, who is counting? The extent of the threat to freshwater fish biodiversity. In ‘Conservation of freshwater fishes’. (Eds GP Closs, M Krkosek, JD Olden) pp. 1–36. (Cambridge University Press) 10.1017/CBO9781139627085.002
Drouineau H, Carter C, Rambonilaza M, Beaufaron G, Bouleau G, Gassiat A, Lambert P, le Floch S, Tétard S, de Oliveira E (2018) River continuity restoration and diadromous fishes: much more than an ecological issue. Environmental Management 61, 671-686.
| Crossref | Google Scholar | PubMed |
El-Sabaawi RW, Frauendorf TC, Marques PS, Mackenzie RA, Manna LR, Mazzoni R, Phillip DAT, Warbanski ML, Zandonà E (2016) Biodiversity and ecosystem risks arising from using guppies to control mosquitoes. Biology Letters 12, 20160590.
| Crossref | Google Scholar |
Godfrey PC, Arthington AH, Pearson RG, Karim F, Wallace J (2017) Fish larvae and recruitment patterns in floodplain lagoons of the Australian Wet Tropics. Marine and Freshwater Research 68(5), 964-979.
| Crossref | Google Scholar |
Guo H, Zhang X, Zhang Y, Tang W, Wu J (2017) Effects of environmental variables on recruitment of Anguilla japonica glass eels in the Yangtze Estuary, China. Fisheries Science 83(3), 333-341.
| Crossref | Google Scholar |
Haryani GS (2021) Sustainable use and conservation of inland water ecosystem in Indonesia: challenge for fisheries management in lake and river ecosystem. In ‘International Conference on the Ocean and Earth Sciences. IOP Conference Series: Earth and Environmental Science, Vol. 789(1)’, 18–20 November 2020, Jakarta Selatan, Indonesia. Paper number 012023. (IOP Publishing Ltd) 10.1088/1755-1315/789/1/012023
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London – B. Biological Sciences 270(1512), 313-321.
| Crossref | Google Scholar |
Heimeier D, Lavery S, Sewell MA (2010) Using DNA barcoding and phylogenetics to identify Antarctic invertebrate larvae: lessons from a large scale study. Marine Genomics 3(3-4), 165-177.
| Crossref | Google Scholar | PubMed |
Herder F, Möhring J, Flury JM, Utama IV, Wantania L, Wowor D, Boneka FB, Stelbrink B, Hilgers L, Schwarzer J, Pfaender J (2022) More non-native fish species than natives, and an invasion of Malawi cichlids, in ancient Lake Poso, Sulawesi, Indonesia. Aquatic Invasions 17(1), 72-91.
| Crossref | Google Scholar |
Hubert N, Delrieu-Trottin E, Irisson J-O, Meyer C, Planes S (2010) Identifying coral reef fish larvae through DNA barcoding: a test case with the families Acanthuridae and Holocentridae. Molecular Phylogenetics and Evolution 55(3), 1195-1203.
| Crossref | Google Scholar | PubMed |
Hubert N, Kadarusman , Wibowo A, Busson F, Caruso D, Sulandari S, Nafiqoh N, Pouyaud L, Rüber L, Avarre J-C, Herder F, Hanner R, Keith P, Hadiaty RK (2015) DNA barcoding Indonesian freshwater fishes: challenges and prospects. DNA Barcodes 3(1), 144-169.
| Crossref | Google Scholar |
Hubert N, Lumbantobing D, Sholihah A, Dahruddin H, Delrieu-Trottin E, Busson F, Sauri S, Hadiaty R, Keith P (2019) Revisiting species boundaries and distribution ranges of Nemacheilus spp. (Cypriniformes: Nemacheilidae) and Rasbora spp. (Cypriniformes: Cyprinidae) in Java, Bali and Lombok through DNA barcodes: implications for conservation in a biodiversity hotspot. Conservation Genetics 20(3), 517-529.
| Crossref | Google Scholar |
Hulley EN, Taylor NDJ, Zarnke AM, Somers CM, Manzon RG, Wilson JY, Boreham DR (2018) DNA barcoding vs. morphological identification of larval fish and embryos in Lake Huron: advantages to a molecular approach. Journal of Great Lakes Research 44(5), 1110-1116.
| Crossref | Google Scholar |
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes 7(4), 544-548.
| Crossref | Google Scholar |
Jo H, Jeppesen E, Ventura M, Buchaca T, Gim J-S, Yoon J-D, Kim D-H, Joo G-J (2019) Responses of fish assemblage structure to large-scale weir construction in riverine ecosystems. Science of The Total Environment 657, 1334-1342.
| Crossref | Google Scholar | PubMed |
Junk WJ, Bayley PB, Sparks RE (1989) The flood-pulse concept in river–floodplain systems. In ‘Proceedings of the International Large River Symposium (LARS)’, 14–21 September 1986, Honey Harbour, ON, Canada. (Ed. DP Dodge) Canadian Special Publication of Fisheries and Aquatic Sciences 106, pp, 110–127. (Fisheries and Oceans Canada: Ottawa, ON, Canada) [In English with abstract in English and French]
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14(6), 587-589.
| Crossref | Google Scholar | PubMed |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12), 1647-1649.
| Crossref | Google Scholar |
Keith P, Lord C, Darhuddin H, Limmon G, Sukmono T, Hadiaty RK, Hubert N (2017) Schismatogobius (Gobiidae) from Indonesia, with description of four new species. Cybium 41(2), 195-211.
| Crossref | Google Scholar |
King J, Brown C (2010) Integrated basin flow assessments: concepts and method development in Africa and South-east Asia. Freshwater Biology 55(1), 127-146.
| Crossref | Google Scholar |
Ko H-L, Wang Y-T, Chiu T-S, Lee M-A, Leu M-Y, Chang K-Z, Chen W-Y, Shao K-T (2013) Evaluating the accuracy of morphological identification of larval fishes by applying DNA barcoding. PLoS ONE 8(1), e53451.
| Crossref | Google Scholar |
Kouhanestani ZM, Ghorbani R, Fujiwara M, Rabaniha M, Amini K, Mahmoodi S (2020) Identification of larval stages of fish in southeastern coastal waters of the Caspian Sea– Golestan Province. Iranian Journal of Fisheries Sciences 19(1), 325-339.
| Crossref | Google Scholar |
Lira NL, Tonello S, Lui RL, Traldi JB, Brandão H, Oliveira C, Blanco DR (2023) Identifying fish eggs and larvae: from classic methodologies to DNA metabarcoding. Molecular Biology Reports 50(2), 1713-1726.
| Crossref | Google Scholar |
Lucas MC, Baras E (2000) Methods for studying spatial behaviour of freshwater fishes in the natural environment. Fish and Fisheries 1(4), 283-316.
| Crossref | Google Scholar |
Mercado-Silva N, Helmus MR, Zanden MJV (2009) The effects of impoundment and non-native species on a river food web in Mexico’s central plateau. River Research and Applications 25(9), 1090-1108.
| Crossref | Google Scholar |
Morgan DL, Gill HS, Maddern MG, Beatty SJ (2004) Distribution and impacts of introduced freshwater fishes in Western Australia. New Zealand Journal of Marine and Freshwater Research 38(3), 511-523.
| Crossref | Google Scholar |
Mueller M, Pander J, Geist J (2011) The effects of weirs on structural stream habitat and biological communities. Journal of Applied Ecology 48(6), 1450-1461.
| Crossref | Google Scholar |
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1), 268-274.
| Crossref | Google Scholar | PubMed |
Park T, Lee G, Shin J, Park J-H, Choi B, Kim D-K, Cha YK (2023) Changes in zooplankton community in response to a shift from lentic to lotic conditions in a regulated river. Ecological Informatics 77, 102236.
| Crossref | Google Scholar |
Rachmatika I, Sjafei DS, Nurcahyadi W (2002) Fish fauna of Gunung Halimun National Park region: additional information on the utilization. Berita Biologi 6(1), 13-24.
| Google Scholar |
Reynalte-Tataje DA, Nakatani K, Fernandes R, Agostinho AA, Bialetzki A (2011) Temporal distribution of ichthyoplankton in the Ivinhema River (Mato Grosso do Sul State/Brazil): influence of environmental variables. Neotropical Ichthyology 9(2), 427-436 [In English with abstract and keywords in English and Portuguese].
| Crossref | Google Scholar |
Reznick D, Butler MJ, IV, Rodd H (2001) Life-history evolution in guppies. VII. The comparative ecology of high- and low-predation environments. The American Naturalist 157(2), 126-140.
| Crossref | Google Scholar |
Ross HA, Murugan S, Sibon Li WL (2008) Testing the reliability of genetic methods of species identification via simulation. Systematic Biology 57(2), 216-230.
| Crossref | Google Scholar |
Schmutz S, Bakken TH, Friedrich T, Greimel F, Harby A, Jungwirth M, Melcher A, Unfer G, Zeiringer B (2015) Response of fish communities to hydrological and morphological alterations in hydropeaking rivers of Austria. River Research and Applications 31(8), 919-930.
| Crossref | Google Scholar |
Soukhaphon A, Baird IG, Hogan ZS (2021) The impacts of hydropower dams in the Mekong River Basin: a review. Water 13(3), 265.
| Crossref | Google Scholar |
Suryaningsih S, Bhagawati D, Sukmaningrum S, Sugiharto S, Puspitasari IGAAR (2020) Freshwater fish diversity in three tributary streams in Serayu Basin, central Java, Indonesia. Biodiversitas Journal of Biological Diversity 21(12), 5811-5817.
| Crossref | Google Scholar |
Teletchea F (2009) Molecular identification methods of fish species: reassessment and possible applications. Reviews in Fish Biology and Fisheries 19, 265-293.
| Crossref | Google Scholar |
Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1), W232-W235.
| Crossref | Google Scholar | PubMed |
Vu AV, Hortle KG, Nguyen DN (2021) Factors driving long term declines in inland fishery yields in the Mekong Delta. Water 13(8), 1005.
| Crossref | Google Scholar |
Ward JV, Stanford JA (1995) Ecological connectivity in alluvial river ecosystems and its disruption by flow regulation. Regulated Rivers: Research & Management 11(1), 105-119.
| Crossref | Google Scholar |
Wibowo A, Wahlberg N, Vasemägi A (2017) DNA barcoding of fish larvae reveals uncharacterised biodiversity in tropical peat swamps of New Guinea, Indonesia. Marine and Freshwater Research 68(6), 1079-1087.
| Crossref | Google Scholar |
Wibowo A, Hubert N, Dahruddin H, Steinke D, Suhaimi RA, Samuel , Atminarso D, Anggraeni DP, Trismawanti I, Baumgartner LJ, Ning N (2021) Assessing temporal patterns and species composition of glass eel (Anguilla spp.) cohorts in sumatra and java using DNA barcodes. Diversity 13, 193.
| Crossref | Google Scholar |
Widén Å, Renöfält BM, Degerman E, Wisaeus D, Jansson R (2021) Let it flow: modeling ecological benefits and hydropower production impacts of banning zero-flow events in a large regulated river system. Science of The Total Environment 783, 147010.
| Crossref | Google Scholar |
Yadav RS, Haq S (2017) Role of fishes in vector-borne disease control in Southeast Asia. In ‘Water and sanitation in the new millennium’. (Eds KJ Nath, VP Sharma) pp. 163–183. (Springer India: New Delhi, India) 10.1007/978-81-322-3745-7_14
Ziegeweid JR, Johnson GD, Krall AL, Fitzpatrick K, Levin SB (2022) Quantifying relations between altered hydrology and fish community responses for streams in Minnesota. Ecological Processes 11, 41.
| Crossref | Google Scholar |