The origins of dengue outbreaks in northern Queensland, Australia, 1990–2017
Alyssa T PykePublic Health Virology Laboratory
Forensic and Scientific Services
Coopers Plains, Qld 4108, Australia
Tel.: +61 7 3096 2865
Fax: +61 7 3096 2878
Email: Alyssa.Pyke@health.qld.gov.au
Microbiology Australia 39(2) 93-95 https://doi.org/10.1071/MA18027
Published: 13 April 2018
Dengue is one of the world’s major infectious mosquito-borne diseases and although not endemic in Australia, is a significant public health concern. Queensland is vulnerable to outbreaks of dengue viruses (DENVs) and indeed, due to endemic populations of the mosquito vector Aedes aeypti, has been the only state since the 1950s to record local transmission. Determining DENV outbreak origins, and monitoring strain movement and diversity greatly assists outbreak management. It also confirms epidemiological links and potentially identifies incursions of rare or highly pathogenic viruses. There have been 73 DENV outbreaks recorded in northern Queensland within the past three decades and it has been the role of Public Health Virology, Department of Health, Queensland Government, to provide DENV genotyping and characterisation to facilitate this essential surveillance. This review summarises the likely origins of the recent northern Queensland outbreaks and describes the complex dynamics of DENV genotypic diversity that have characterised local transmission events.
Boasting tropical and subtropical climates, Queensland attracts more than two million overseas visitors annually1. The increased frequency and affordability of travel and expanded business and trade have contributed to the influx of travellers to Queensland, including visitors and returning Queensland residents coming from Southeast Asia, the western Pacific and other dengue virus (DENV) endemic regions. Populations of the primary DENV mosquito vector Aedes aegypti are present on mainland Queensland and the Torres Strait islands2. Another DENV vector, Ae. albopictus, is currently restricted to the Torres Strait islands3. Thus, Queensland is prone to DENV outbreaks and other arthropod-borne virus disease threats, such as chikungunya and Zika viruses4,5.
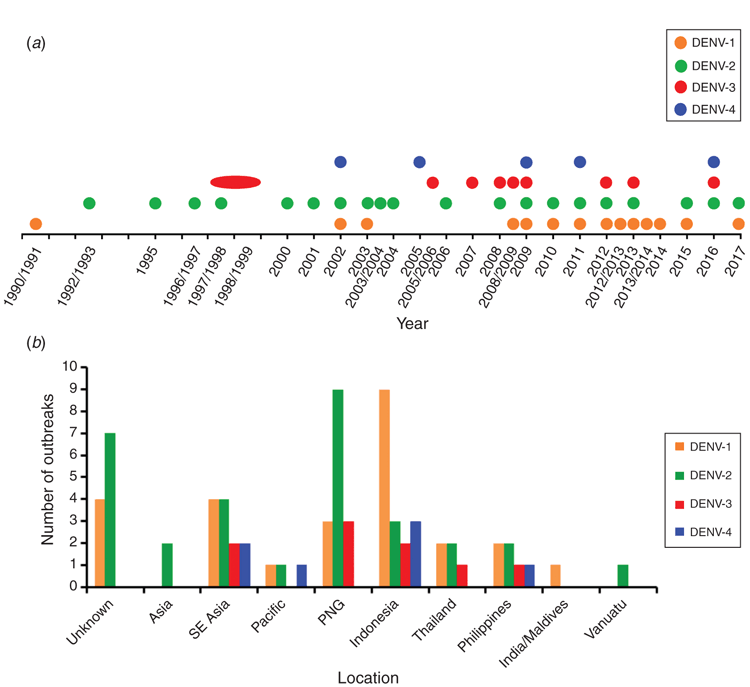
At least 73 DENV outbreaks, each involving one or more locally acquired case(s), have been recorded in northern Queensland between 1990 and 20176–14 (Dianne Brookes, Tropical Public Health Unit (TPHU) Cairns, and Jan Humphreys, TPHU Townsville, unpublished data) (Figure 1a). Whilst all four DENV serotypes (DENV 1–4) have caused outbreaks (DENV-1, 36%; DENV-2, 42%; DENV-3, 12%; DENV-4, 10%), DENV-1 and DENV-2 account for almost 80% of outbreaks. Major outbreak epicentres included the Torres Strait islands (northernmost outbreak location), Cairns, Townsville, Port Douglas, Mossman, Kuranda, Mareeba, Innisfail, Tully, Ingham and Charters Towers (furthest southern/western outbreak location). Recent outbreak trends for the period between 1990 and 2016, showed a steady increase in the recorded number of outbreaks, despite variable numbers of locally acquired cases during the same period and a recent decline in cases between 2011 and 20168. Determining the origin and genetic relatedness of DENV strains plays a key role in disease mitigation strategies, by affirming epidemiological links and providing early warning of sustained transmission or, importantly, endemicity, if it was to occur.
For most DENV outbreaks, index cases are not known and local transmission is normally identified after laboratory confirmation of DENV positive patient(s) who have no history of travel abroad. Therefore, the determination of outbreak origins has become increasingly reliant on viral nucleotide sequencing and phylogenetic molecular techniques. Nucleotide sequencing and phylogenetic analysis of complete DENV envelope (E) genes was used to establish the possible geographical origins of Australian outbreak strains and compare their genetic diversity with other imported and globally circulating DENVs. Figure 1b summarises the number of outbreaks between 1990 and 2017 and their most likely overseas geographical sources based on phylogenetic analyses. The data include primary outbreaks initiated by viraemic international travellers and secondary outbreaks likely resulting from further spread of the same virus strain(s) to new locations (for example, Cairns to Townsville). For the current report, secondary outbreaks were also assigned the likely overseas source to highlight the impact of respective overseas incursions on transmission and disease.
Likely origins of outbreaks were determined following sequence alignment and phylogenetic tree analysis as previously described8,9. Outbreak strains were considered to have been sourced from a particular region after demonstrating very high percentage sequence identities (≥99%) and evolutionary relatedness with the strain identified and sequenced from the index case (where available) or clusters of strains (two or more) which were themselves closely related and recently circulating in that region. Specific country categories included Papua New Guinea (PNG), Indonesia, Thailand, the Philippines, Vanuatu and India/Maldives. For some outbreaks, a broader regional classification (Asia, Southeast Asia and the Pacific) was assigned if related phylogenetic clusters displayed an ambiguous geographical source (contained sequences from several countries of that region) or the outbreak strain was only closely related to one other DENV strain. Unfortunately, the origin of 11 outbreaks (4 DENV-1 and 7 DENV-2) could not be determined due to absence of isolates or DENV PCR positive patient samples for sequencing.
The largest number of outbreaks by country involved imported strains from Indonesia (n = 17; 23%). Indonesia has been shown previously to be a major source of imported DENVs into Queensland, Australia9, and likely reflects the high frequency of travellers from this country, in particular, Bali. Within the last seven years, the annual number of visitors who travelled from Indonesia to Queensland was ≈20 0001. PNG accounted for the second highest number of outbreaks (n = 15; 21%) and remains a major source of imported DENVs into northern Queensland8. Collectively, Southeast Asia (including Indonesia, the Philippines and Thailand) was responsible for 40 outbreaks (55%) and continues to be the highest contributor of outbreaks, due to all four DENV serotypes9.
In addition to the co-circulation of multiple serotypes, some concurrent or closely successive outbreaks were caused by a single DENV serotype, and it was only after performing phylogenetic analysis, that involvement of one or more viral strain(s) could be determined. Occasionally, this further assisted the identification of secondary outbreaks in the absence of clear epidemiological links. In 2003–2004, three unrelated strains of DENV-2 (two from PNG and one from Thailand) were responsible for outbreaks in north Queensland resulting in ≈890 cases, including two haemorrhagic cases and one death7. Phylogenetic analysis revealed that three separate DENV-2 strains were responsible for outbreaks in (1) Cairns, early 2003, and two later subsequent outbreaks in Townsville, 2003, (2) Cairns, late 2003, and (3) the Torres Strait islands, late 2003, followed by an outbreak in Cairns, early 20047. Similarly, in 2012–2013, four different DENV-1 strains from Southeast Asia (including one from Thailand and two from Bali) were responsible for separate outbreaks in (1) Cairns/Townsville/Ingham, (2) Townsville, (3) Innisfail and (4) Cairns.
Sustained transmission of DENV strains over long, uninterrupted periods increases the risk of endemicity. Although northern Queensland outbreaks have not resulted in endemicity to date, several prolonged outbreak events, extending from summer through winter have occurred, including DENV-3 in 1997–1999 (70 weeks duration, 498 cases) originating from Thailand11, DENV-2 in 2003–2004 (69 weeks duration, ≈500 cases) originating from PNG7 and DENV-1 in 2012–2013 (29 weeks duration, ≈170 cases) originating from Thailand (Dianne Brookes, TPHU Cairns, and Jan Humphreys, TPHU Townsville, unpublished data).
In the absence of a suitable vaccine, concerted mosquito control aimed at supressing Ae. aegypti and Ae. albopictus populations is the primary strategy for counteracting the DENV threat in northern Queensland. Biological approaches such as the release of Wolbachia infected Ae. aegypti are also being trialled in several high risk areas to potentially reduce transmission15. Importantly, constant vigilance and surveillance to detect DENV introduction and identify potential geographical sources of strains is crucial for early case identification, accurate monitoring of strain dissemination and disease epidemiology. These factors also ensure that limited mosquito control resources are deployed where they can have the greatest impact, thereby further confining DENV outbreaks in northern Australia.
Acknowledgements
Staff from the Cairns and Townsville Tropical Public Health Units, private and public Queensland hospitals, Sullivan and Nicolaides and Queensland Medical Laboratory private pathology laboratories and the Forensic and Scientific Services (FSS) Public Health Virology Laboratory are gratefully acknowledged for contributing to patient diagnosis, virus genotyping and collation of outbreak data.
References
[1] Tourism Research Australia (2018) International Visitor Survey, extracted by Andrew Wynne-Jones, Queensland Treasury and Coralie Palmeri, Tourism Research Australia. https://www.tra.gov.au/Research/Regional-tourism/local-government-area-profiles (accessed January and March 2018).[2] Beebe, N.W. et al. (2009) Australia’s dengue risk driven by human adaptation to climate change. PLoS Negl. Trop. Dis. 3, e429.
| Australia’s dengue risk driven by human adaptation to climate change.Crossref | GoogleScholarGoogle Scholar |
[3] van den Hurk, A.F. et al. (2016) Ten years of the Tiger: Aedes albopictus presence in Australia since its discovery in the Torres Strait in 2005. One Health 2, 19–24.
| Ten years of the Tiger: Aedes albopictus presence in Australia since its discovery in the Torres Strait in 2005.Crossref | GoogleScholarGoogle Scholar |
[4] Hall-Mendelin, S. et al. (2016) Assessment of local mosquito species incriminates Aedes aegypti as the potential vector of Zika virus in Australia. PLoS Negl. Trop. Dis. 10, e0004959.
| Assessment of local mosquito species incriminates Aedes aegypti as the potential vector of Zika virus in Australia.Crossref | GoogleScholarGoogle Scholar |
[5] van den Hurk, A.F. et al. (2010) Vector competence of Australian mosquitoes for chikungunya virus. Vector Borne Zoonotic Dis. 10, 489–495.
| Vector competence of Australian mosquitoes for chikungunya virus.Crossref | GoogleScholarGoogle Scholar |
[6] Hanna, J.N. and Ritchie, S.A. (2009) Outbreaks of dengue in north Queensland, 1990–2008. Commun. Dis. Intell. 33, 32–33.
[7] Hanna, J.N. et al. (2006) Multiple outbreaks of dengue serotype 2 in north Queensland, 2003/04. Aust. N. Z. J. Public Health 30, 220–225.
| Multiple outbreaks of dengue serotype 2 in north Queensland, 2003/04.Crossref | GoogleScholarGoogle Scholar |
[8] Moore, P.R. et al. (2017) Dengue viruses in Papua New Guinea: evidence of endemicity and phylogenetic variation, including the evolution of new genetic lineages. Emerg. Microbes Infect. 6, e114.
| Dengue viruses in Papua New Guinea: evidence of endemicity and phylogenetic variation, including the evolution of new genetic lineages.Crossref | GoogleScholarGoogle Scholar |
[9] Warrilow, D. et al. (2012) Sources of dengue viruses imported into Queensland, Australia, 2002–2010. Emerg. Infect. Dis. 18, 1850–1857.
| Sources of dengue viruses imported into Queensland, Australia, 2002–2010.Crossref | GoogleScholarGoogle Scholar |
[10] Hanna, J.N. et al. (2009) Dengue in north Queensland, 2005–2008. Commun. Dis. Intell. 33, 198–203.
[11] Hanna, J.N. et al. (2001) An epidemic of dengue 3 in far north Queensland, 1997–1999. Med. J. Aust. 174, 178–182.
| 1:STN:280:DC%2BD3M7nsFyisQ%3D%3D&md5=334bd4cf7e8029588f11832a444c0b16CAS |
[12] Hanna, J.N. et al. (1998) Two contiguous outbreaks of dengue type 2 in north Queensland. Med. J. Aust. 168, 221–225.
| 1:STN:280:DyaK1c3gtVOhsg%3D%3D&md5=25429058a42591a250326c777104b12fCAS |
[13] Hanna, J.N. et al. (2003) Dengue in north Queensland, 2002. Commun. Dis. Intell. 27, 384–389.
[14] Ritchie, S.A. et al. (2013) An explosive epidemic of DENV-3 in Cairns, Australia. PLoS One 8, e68137.
| An explosive epidemic of DENV-3 in Cairns, Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1Shu7fJ&md5=634d2fead9e8a25d1356894f7f8ed42eCAS |
[15] Eliminate Dengue Australia Program (2018) Eliminate Dengue. http://www.eliminatedengue.com/australia (accessed February 2018).
Biography
Dr Alyssa Pyke is a Supervising Research Virologist at the Public Health Virology Laboratory, Forensic and Scientific Services, Queensland Health. She is primarily involved in the detection, surveillance and characterisation of viruses of public health significance. Her research interests include diagnostic assay development, utilisation of next generation sequencing for molecular epidemiology, characterisation of rare viruses and investigations into arbovirus emergence and evolution.