Newly discovered mosquito viruses help control vector-borne viral diseases
Roy A Hall A and Jody Hobson-Peters BA Australian Infectious Diseases Research Centre
School of Chemistry and Molecular Biosciences
The University of Queensland
St Lucia, Qld 4072, Australia
Email: roy.hall@uq.edu.au
B Australian Infectious Diseases Research Centre
School of Chemistry and Molecular Biosciences
The University of Queensland
St Lucia, Qld 4072, Australia
Email: j.peters2@uq.edu.au
Microbiology Australia 39(2) 72-75 https://doi.org/10.1071/MA18020
Published: 13 April 2018
Many well-known mosquito-borne viruses such as dengue, Zika, West Nile, chikungunya and Ross River viruses can be transmitted to vertebrates and are associated with disease in man or animals. However, the use of deep sequencing and other open-minded approaches to detect viruses in mosquitoes have uncovered many new RNA viruses, most of which do not infect vertebrates. The discovery of these ‘insect-specific’ viruses (ISVs) has redefined the mosquito virome and prompted the lines of viral taxonomic classification to be redrawn1,2. Despite their benign phenotype, ISVs have become a hot topic of research, with recent studies indicating they have significant application for biotechnology.
The main focus of our lab is the study of new and emerging mosquito-borne viruses. For most of the past decade we and our collaborators have developed a comprehensive system for high throughput virus detection and isolation from mosquito and vertebrate samples. This has enabled the discovery of many new viruses and detection of known viruses occurring in new ecological or pathological contexts. We have also focussed on the development of novel research tools and reagents to characterise these viruses both in vitro and in vivo.
To conduct investigations into the biodiversity of viruses in Australian mosquito populations, we have had access to extensive archival collections of mosquito pools collected from different parts of Australia over several decades. These collections were part of previous targeted research projects or routine surveillance operations and were pivotal to the success of our virus discovery program. Another key to our success was the development of a sequence-independent system to detect and isolate new and known viruses in a high throughput manner. This was based on a novel set of monoclonal antibodies we generated specific to double-stranded RNA (dsRNA), which have the crucial ability to recognise the replicative dsRNA intermediates produced by most RNA viruses during growth in cell culture. These antibodies, also known as ‘MAVRIC’ (monoclonal antibodies to viral replicative intermediates in cells), are used in ELISA to detect viral replication in C6/36 mosquito cells in 96-well plates inoculated with mosquito samples3. This allows us to target the MAVRIC-positive cultures for viral isolation and amplification by generic viral RT-PCRs or deep sequencing to identify the viral agent.
To date, the work of several postdocs as well as PhD and honours students in the lab has resulted in the detection, isolation and characterisation of more than 20 new arthropod-borne viruses. These new viruses represent at least nine viral taxa, including flaviviruses, bunyaviruses, mesoniviruses, negeviruses, reoviruses, iflaviruses, nodaviruses, birnaviruses and totiviruses4–12. It is interesting to note that only one of the newly discovered viruses was able to infect vertebrate cells, albeit in a highly restricted fashion4. The high yield of these new insect-specific viruses in our studies likely reflects the fact that previous approaches for virus discovery and surveillance have relied on the use of vertebrate systems (mice or cell lines) for the detection and isolation of mosquito-borne viruses. Whilst this was effective in the discovery of many true arboviruses that cycle between mosquitoes and vertebrates, such methods preclude the detection of insect-specific viruses.
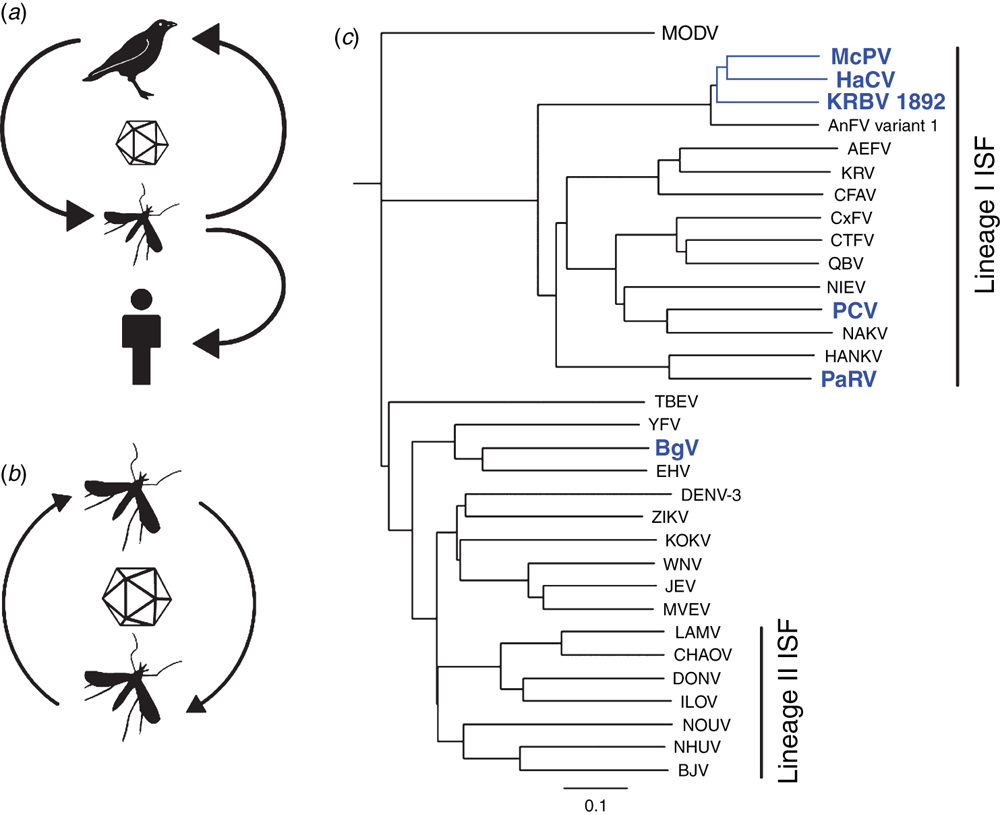
Most of our efforts to characterise these new viruses have focussed on the insect-specific flaviviruses (ISFs). While ISFs share the same genome structure and basic replication strategy as flavivirus pathogens such as West Nile (WNV), Zika (ZIKV) and dengue (DENV) viruses, they do not infect or replicate in vertebrates (Figure 1a, b). Phylogenetic analysis of ISFs also group them into two distinct genetic clusters – referred to as Lineage I ISFs and Lineage II ISFs (reviewed in1 – see Figure 1c). The Lineage I ISFs are the most genetically divergent and are thought to represent the ancestors of all flaviviruses. This supports the hypothesis that all arboviruses originally evolved in arthropods2. Lineage II ISFs on the other hand are genetically much more closely related to the pathogenic flaviviruses and are hypothesised to have recently evolved from a vertebrate-infecting ancestor. This also provides support for convergent evolution amongst ISFs.
The inability to replicate in a vertebrate host indicates that ISFs utilise a form of vertical transmission, a process that has been demonstrated in the laboratory for a number of ISF species13,14 (Figure 1c). Our own studies with Parramatta River virus (PaRV), a Lineage I ISF isolated from Aedes vigilax in Sydney, revealed that a high proportion of both male and female progeny of wild-caught female mosquitoes that were hatched and reared in the laboratory were infected with PaRV (unpublished data). Just how the virus infects progeny mosquitoes via the egg has not been determined.
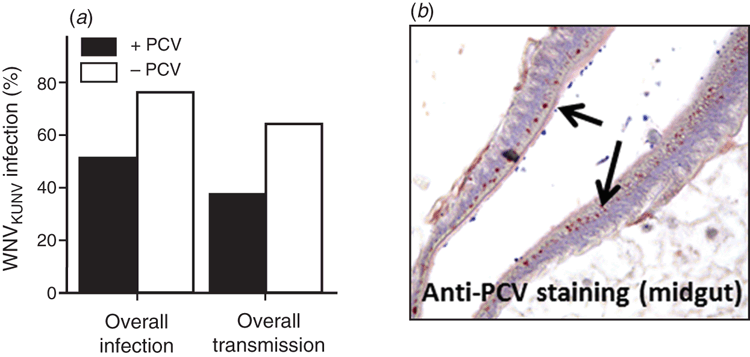
Efficient vertical transmission by ISFs can result in a very high frequency of infected mosquitoes in some populations. This has been shown to reach 80–100% in some studies6,14. Curiously, a high prevalence of persistent ISF infection in mosquito populations may have a significant effect on the transmission of flavivirus pathogens such as WNV or dengue. Indeed, laboratory studies by us and others have shown that female Culex mosquitoes previously infected (naturally or artificially) with Lineage I or Lineage II ISFs reduced their susceptibility to infection by WNV and their ability to transmit this virus, likely due to the apparent localisation of ISF replication to the cells of the mosquito mid-gut15,16 (Figure 2). This suggests that ISFs may naturally regulate the transmission of pathogens in some mosquito populations and may present an opportunity to develop novel strategies to reduce the transmission of mosquito-borne viral disease.
|
To understand why ISFs do not replicate in vertebrate cells, we developed a series of research tools to identify the stages of the cellular replication cycle at which restriction occurs. These included monoclonal antibodies to detect the viral proteins of ISFs produced during replication, and infectious DNA-clones of these viruses to identify viral factors associated with host restriction6,9,17,18. These infectious DNAs have enabled us to replace different parts of the ISF genome with the corresponding region of West Nile virus, a flavivirus that successfully replicates in most vertebrate cell types. These chimeric viruses have revealed that the structural genes that code for the virion envelope proteins of ISFs are unable to facilitate entry of the virus to vertebrate cells, while components in the remainder of the genome (non-structural proteins and untranslated regions of the positive strand RNA genome) render ISFs incapable of initiating replication in the cytoplasm of vertebrate cells.
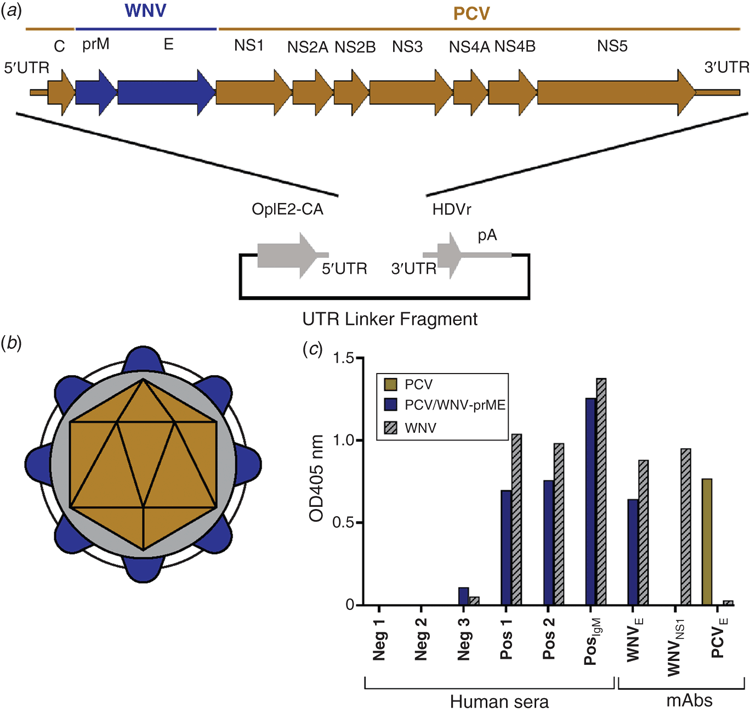
Our success in producing chimeric viruses between an ISF (PCV) and a pathogenic flavivirus (WNV) led us to express the immunogenic antigens of WNV and other pathogenic flaviviruses to develop a new platform for the safe and simple production of diagnostic antigens and vaccine candidates. Using PCV as the genetic backbone, we were able to construct viable chimeric viruses that expressed the prM and E virion proteins of WNV, ZIKV or DENV 218 (Figure 3a, b; unpublished data) that are antigenically authentic and suitable as diagnostic antigens (Figure 3c). Subsequently, we have identified other ISF species that can be also used for this purpose. Importantly, the chimeric viruses exhibit the host restriction phenotype of the parental ISF and do not replicate in vertebrate cells. They can also be grown to high titre in mosquito cell culture, often to orders of magnitude greater than the parental pathogenic virus.
|
Future directions
We are currently elucidating the precise mechanisms involved in the transmission of ISFs and the ecological context in which this occurs. Elucidating the molecular basis of their host restriction and how this relates to the evolution of different ISF lineages is also another interesting facet of our research. While the application of ISFs to develop recombinant platforms for safe diagnostics and vaccines provides exciting potential for biotechnology, we are also intrigued by the apparent regulation of pathogen transmission in mosquito populations that carry ISFs and the possibilities of exploiting this phenomenon to control the transmission of flavivirus diseases.
Acknowledgements
We acknowledge the many staff and students in the School of Chemistry and Molecular Biosciences at UQ and in Public Health Virology at Queensland Health who have contributed to this research and to the many collaborators who have provided arthropod and clinical samples that have enabled us to undertake the work. Special thanks to Agathe Colmant for preparing the phylogenetic tree, Jessica Harrison and Greg Sullivan for editing the manuscript and Tobias Hall for the schematic artwork.
References
[1] Hall, R.A. et al. (2016) Commensal viruses of mosquitoes: host restriction, transmission and interactions with arboviral pathogens. Evol. Bioinform. Online 12, 35–44.[2] Shi, M. et al. (2016) Redefining the invertebrate RNA virosphere. Nature 540, 539–543.
| Redefining the invertebrate RNA virosphere.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhvFelsbfM&md5=9143d7221da4e190b501b318cf1201cfCAS |
[3] O’Brien, C.A. et al. (2015) Viral RNA intermediates as targets for detection and discovery of novel and emerging mosquito-borne viruses. PLoS Negl. Trop. Dis. 9, e0003629.
[4] Colmant, A.M.G. et al. (2016) A newly discovered flavivirus in the yellow fever group displays restricted replication in vertebrates. J. Gen. Virol. 97, 1087–1093.
| A newly discovered flavivirus in the yellow fever group displays restricted replication in vertebrates.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsFKiurnK&md5=19e31c2f46c3d92b7f6607de7891bd7aCAS |
[5] Colmant, A.M.G. et al. (2017) Discovery of new orbiviruses and totivirus from Anopheles mosquitoes in Eastern Australia. Arch. Virol. 162, 3529–3534.
| Discovery of new orbiviruses and totivirus from Anopheles mosquitoes in Eastern Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXht1yltLzO&md5=ef2c7996570aa30ddaacee4f6a41dbe6CAS |
[6] Colmant, A.M.G. et al. (2017) A new clade of insect-specific flaviviruses from Australian Anopheles mosquitoes displays species-specific host restriction. MSphere 2, e00262-17.
| A new clade of insect-specific flaviviruses from Australian Anopheles mosquitoes displays species-specific host restriction.Crossref | GoogleScholarGoogle Scholar |
[7] Harrison, J.J. et al. (2016) A new orbivirus isolated from mosquitoes in North-Western Australia shows antigenic and genetic similarity to Corriparta virus but does not replicate in vertebrate cells. Viruses 8, 141–156.
| A new orbivirus isolated from mosquitoes in North-Western Australia shows antigenic and genetic similarity to Corriparta virus but does not replicate in vertebrate cells.Crossref | GoogleScholarGoogle Scholar |
[8] Hobson-Peters, J. et al. (2016) Discovery and characterisation of a new insect-specific bunyavirus from Culex mosquitoes captured in northern Australia. Virology 489, 269–281.
| Discovery and characterisation of a new insect-specific bunyavirus from Culex mosquitoes captured in northern Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhvFClsbjO&md5=12347127345f55efdf5ebdc3466f047fCAS |
[9] Hobson-Peters, J. et al. (2013) A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One 8, e56534.
| A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjvV2hsL0%3D&md5=8e0b6d71e1bb1dc8676a20358bf4b567CAS |
[10] McLean, B.J. et al. (2015) A novel insect-specific flavivirus replicates only in Aedes-derived cells and persists at high prevalence in wild Aedes vigilax populations in Sydney, Australia. Virology 486, 272–283.
| A novel insect-specific flavivirus replicates only in Aedes-derived cells and persists at high prevalence in wild Aedes vigilax populations in Sydney, Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtleksrjN&md5=c77eb38ae4b97607d0ac968ac2e60b75CAS |
[11] O’Brien, C.A. et al. (2017) Discovery and characterisation of Castlerea virus, a new species of negevirus isolated in Australia. Evol. Bioinform. Online 13, .
| Discovery and characterisation of Castlerea virus, a new species of negevirus isolated in Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC1cXjsFCitLw%3D&md5=11265a6cfd266cee2a6dfa6ec42cdd88CAS |
[12] Warrilow, D. et al. (2014) A new species of mesonivirus from the Northern Territory, Australia. PLoS One 9, e91103.
| A new species of mesonivirus from the Northern Territory, Australia.Crossref | GoogleScholarGoogle Scholar |
[13] Blitvich, B.J. et al. (2015) Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organisation. Viruses 7, 1927–1959.
| Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organisation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXotFKht7s%3D&md5=e1e38f2de94839bfb97e04aa5160a412CAS |
[14] Bolling, B.G. et al. (2012) Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colong and effects of co-infection on vector competence for West Nile virus. Virology 427, 90–97.
| Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colong and effects of co-infection on vector competence for West Nile virus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xkt1Gjsr0%3D&md5=0fd9f750948f7267dd17b3037b2a743aCAS |
[15] Goenaga, S. et al. (2015) Potential for co-infection of a mosquito-specific flavivirus, Nhumirim virus, to block West Nile virus transmission in mosquitoes. Viruses 7, 5801–5812.
| Potential for co-infection of a mosquito-specific flavivirus, Nhumirim virus, to block West Nile virus transmission in mosquitoes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhtFOntbfP&md5=efa308ffc17409229e91ac47bf08df19CAS |
[16] Hall-Mendelin, S. et al. (2016) The insect-specific Palm Creek virus modulates West Nile infection in and transmission by Australian mosquitoes. Parasit. Vectors 9, 414–424.
| The insect-specific Palm Creek virus modulates West Nile infection in and transmission by Australian mosquitoes.Crossref | GoogleScholarGoogle Scholar |
[17] Junglen, S. et al. (2017) Host range restriction of insect-specific flaviviruses occurs at several levels of the viral life cycle. MSphere 2, e00375-16.
| Host range restriction of insect-specific flaviviruses occurs at several levels of the viral life cycle.Crossref | GoogleScholarGoogle Scholar |
[18] Piyasena, T.B. et al. (2017) Infectious DNAs derived from insect-specific flavivirus genomes enable identification of pre- and post-entry mechanisms of host restriction in vertebrate cells. Sci. Rep. 7, 2940.
| Infectious DNAs derived from insect-specific flavivirus genomes enable identification of pre- and post-entry mechanisms of host restriction in vertebrate cells.Crossref | GoogleScholarGoogle Scholar |
Biographies
Professor Roy Hall is a specialist in vector-borne virology at the University of Queensland. His research explores emerging mosquito-borne viruses with a focus on their pathogenesis and the development of novel vaccine and diagnostic platforms. His work has led to the design and development of novel diagnostic assays and vaccine candidates and the discovery of several new mosquito-borne viruses.
Dr Jody Hobson-Peters is a virologist at the University of Queensland specialising in mosquito-borne virus discovery and the development of novel diagnostic assays. Following almost a decade working in industry in the development and commercialisation of rapid point-of-care assays, her most recent research interests have culminated in a greater understanding of the mosquito virome, producing an extensive suite of monoclonal antibodies to novel mosquito-borne viruses and the optimisation of safe and authentic viral protein production for next-generation mosquito-borne virus vaccines and diagnostics.