OzFoodNet into the future: the rapid evolution of foodborne disease surveillance in Australia
Ben Polkinghorne A , Anthony Draper B , Michelle Harlock C and Robyn Leader DA OzFoodNet Coordinating Epidemiologist, Office of Health Protection, Australian Government Department of Health. Corresponding author: Tel: +61 2 6289 1831, Email: ben.polkinghorne@health.gov.au
B OzFoodNet Epidemiologist, Centre for Disease Control, Department of Health, Northern Territory Government. Tel: +61 8 8922 7635, Email: anthony.draper@nt.gov.au
C OzFoodNet Epidemiologist, Communicable Disease Prevention Unit, Department of Health and Human Services, Tasmanian Government. Tel: +61 3 6166 0667, Email: michelle.harlock@dhhs.tas.gov.au
D Assistant Director, Office of Health Protection, Australian Government Department of Health. Tel: +61 2 6289 2750, Email: robyn.leader@health.gov.au
Microbiology Australia 38(4) 179-183 https://doi.org/10.1071/MA17063
Published: 2 November 2017
OzFoodNet is Australia’s national enhanced foodborne disease surveillance network. OzFoodNet is currently evolving in order to meet the most significant challenges faced since it commenced in 2000: the transition to culture independent diagnostic tests and the introduction of whole genome sequencing for typing of enteric pathogens. This has changed the nature of foodborne disease surveillance and outbreak investigation in Australia.
OzFoodNet is Australia’s national enhanced foodborne disease surveillance network, with foodborne disease epidemiologists in each Australian state and territory and the Australian Department of Health and representatives from Food Standards Australia New Zealand, Australian Government Department of Agriculture and Water Resources, the Public Health Laboratory Network (PHLN) and the National Centre for Epidemiology and Population Health1. OzFoodNet is over 15 years old and continues to successfully conduct surveillance and respond to outbreaks of foodborne illness. Relationships have been crucial to this success, and triannual face-to-face meetings have been the cornerstone of OzFoodNet’s collaboration. OzFoodNet celebrated its 50th face-to-face meeting in Canberra, in November 2016 (Figure 1). This meeting involved reflection on past achievements, but as always was a working meeting, with Australian and international experts present to discuss current challenges faced by OzFoodNet and to assist in planning for its future.

|
The nature of foodborne disease surveillance and outbreak investigation in Australia has changed radically in the last four years. In 2013, a number of diagnostic laboratories began implementing culture independent diagnostic testing (CIDT) in the form of multiplex polymerase chain reaction (PCR) tests that target up to 10 enteric pathogens per test. These PCR tests are typically more sensitive than traditional culture as they detect not only viable organisms but also the nucleic acid of organisms that may not have been able to be cultured by traditional culture methods. This provides potential benefits for the clinician by providing a more rapid diagnosis and has also increased notifications of enteric pathogens due to increased sensitivity. However, these PCR tests do not allow for further characterisation of pathogens, which OzFoodNet epidemiologists rely on to detect clusters and investigate potential outbreaks, and preclude antimicrobial sensitivity testing.
A challenge described during OzFoodNet’s first decade was the diversity of subtyping methods available for Salmonella Typhimurium, which made the detection and investigation of multi-jurisdictional outbreaks (MJO) very difficult1. Multiple-locus variable number tandem repeat analysis (MLVA) is now established nationally and there is standardised methodology and nomenclature across the country that assists in rapid cluster detection. However, this technique is not readily comparable internationally, which in the context of an increasingly complex global food chain is problematic. Additionally, whole genome sequencing (WGS) for foodborne pathogens has emerged rapidly in Australia and will soon render MLVA and other routine subtyping methods obsolete.
Culture independent diagnostic testing
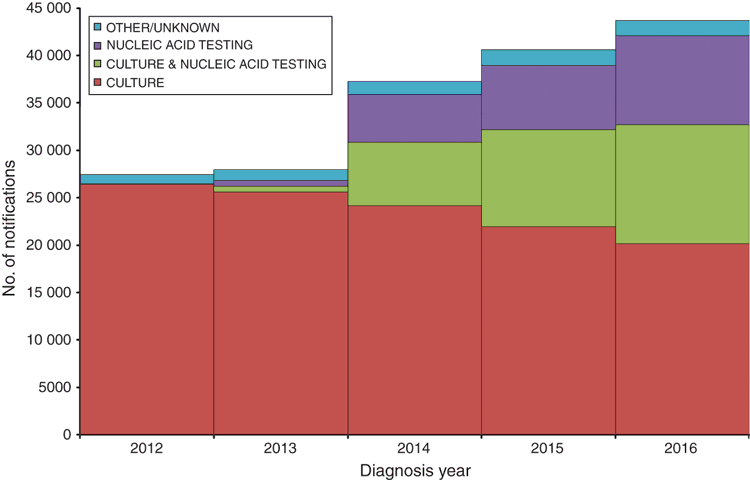
CIDT was a major theme discussed at the 50th OzFoodNet face-to-face meeting. There has been a 59% increase in notifications of the three most common bacterial enteric diseases (salmonellosis, campylobacteriosis, and shigellosis) from 2012 to 2016 (Figure 2). The increase in culture independent notifications since the introduction of the multiplex PCR panels in late 2013 has resulted in CIDT-only notifications accounting for 22% of all notifications for these diseases in 2016 (9402/43 646). The majority of these notifications were for campylobacteriosis (29%, 7077/24 166) and shigellosis (28%, 397/1407). The PCR target for Shigella is shared with enteroinvasive Escherichia coli, which is not notifiable, complicating case follow up. However, 11% of salmonellosis notifications were by CIDT only (1928/18 073) representing a loss of typing information used for public health surveillance and outbreak detection. A positive development has been the increasing number of notifications generated by a combination of CIDT and culture methods, usually a result of pathology labs initially screening by CIDT and then retrospectively culturing all positive samples (‘reflex testing’) for referral to public health laboratories for the purpose of public health surveillance. CIDT and culture testing was performed for 29% of notifications for these diseases in 2016, with 31% of salmonellosis notifications (5663/18 073) being made this way. This is of great benefit to public health but may not be economically viable for laboratories in the long term.
Whole genome sequencing
OzFoodNet has performed enhanced surveillance on all notified cases of invasive listeriosis nationally since 2010 through its National Enhanced Listeriosis Surveillance System (NELSS). All Listeria monocytogenes isolates from these cases were subtyped using pulsed field gel electrophoresis (PFGE) performed at the Microbiological Diagnostic Unit Public Health Laboratory (MDU) in Melbourne, Victoria. PFGE is a slow, technically demanding and resource intensive process2, so several other less discriminatory but rapid subtyping methods (binary typing, molecular serotyping and MLVA) were performed at MDU and other enteric reference laboratories to assist with rapid cluster detection. Since 1 July 2015, MDU began to routinely perform WGS in parallel with PFGE and after a 12 month trial, WGS was accepted as the gold standard for subtyping of invasive listeriosis cases in Australia. This method provides unparalleled resolution for detecting clusters of this relatively rare but frequently severe disease2. The median number of days from notification to a PFGE result being entered into NELSS was 51 days, based on 2010–2013 data3. This has reduced to a matter of a few weeks under routine WGS, with MDU supplying a fortnightly report on all recent human and environmental isolates.
WGS was instrumental in detecting and investigating a multi-jurisdictional outbreak of listeriosis in 2016. Seven cases of invasive listeriosis across four states were found to be highly related to each other and also to food and environmental specimens from three supermarket delicatessens. An eighth probable case was also potentially linked to the outbreak cases by WGS, but also highly related to an environmental sample from a smallgoods manufacturer. The epidemiological investigation was unable to detect a particular product common to all cases, so WGS was integral in identifying that these isolates were all closely related and it is unlikely that OzFoodNet would have linked these cases together without the superior discriminating ability of WGS4.
WGS has also facilitated direct comparisons with international data. This is essential as the global food chain becomes increasingly complex. In 2015 a single case of invasive listeriosis in Queensland was not found to be closely linked to any others in Australia by WGS or epidemiologically; however, on sharing the sequence with international databases it was found to be almost identical to a cluster of cases in the United States of America (USA). These USA cases were associated with contaminated stone fruit that had also been imported to Australia and the Australian case indeed recalled consuming imported peaches. This led to a recall of the imported fruit5.
The complexity of WGS and relatedness of L. monocytogenes isolates has become something that OzFoodNet epidemiologists have had to quickly learn and adapt to, with clusters now being detected initially by laboratory bioinformatics systems, bioinformaticians and laboratory scientists rather than epidemiologists interpreting a traditional laboratory result. Epidemiological information remains crucial in terms of validating clusters and conducting investigations. The evolution of these techniques and the surveillance systems that need to be developed has led to scientists and epidemiologists collaborating more closely together than ever before.
Whole genome sequencing in recent multi-jurisdictional outbreak investigations
Each year OzFoodNet typically investigates several clusters and outbreaks that cross state and territory or international borders. When OzFoodNet deems an outbreak to be of significant size, complexity, or severity to warrant a coordinated national epidemiological investigation, it recommends to the Communicable Diseases Network Australia (CDNA), the national peak body for the prevention and control of communicable diseases, that a multi-jurisdictional epidemiological outbreak investigation (MJOI) commence. These investigations occur under the Guidelines for the epidemiological investigation of multi-jurisdictional outbreaks that are potentially foodborne (the Guidelines). Under the Guidelines, OzFoodNet conducts the epidemiological investigation under a single Lead Epidemiologist and in close collaboration with the human laboratory and environmental investigations.
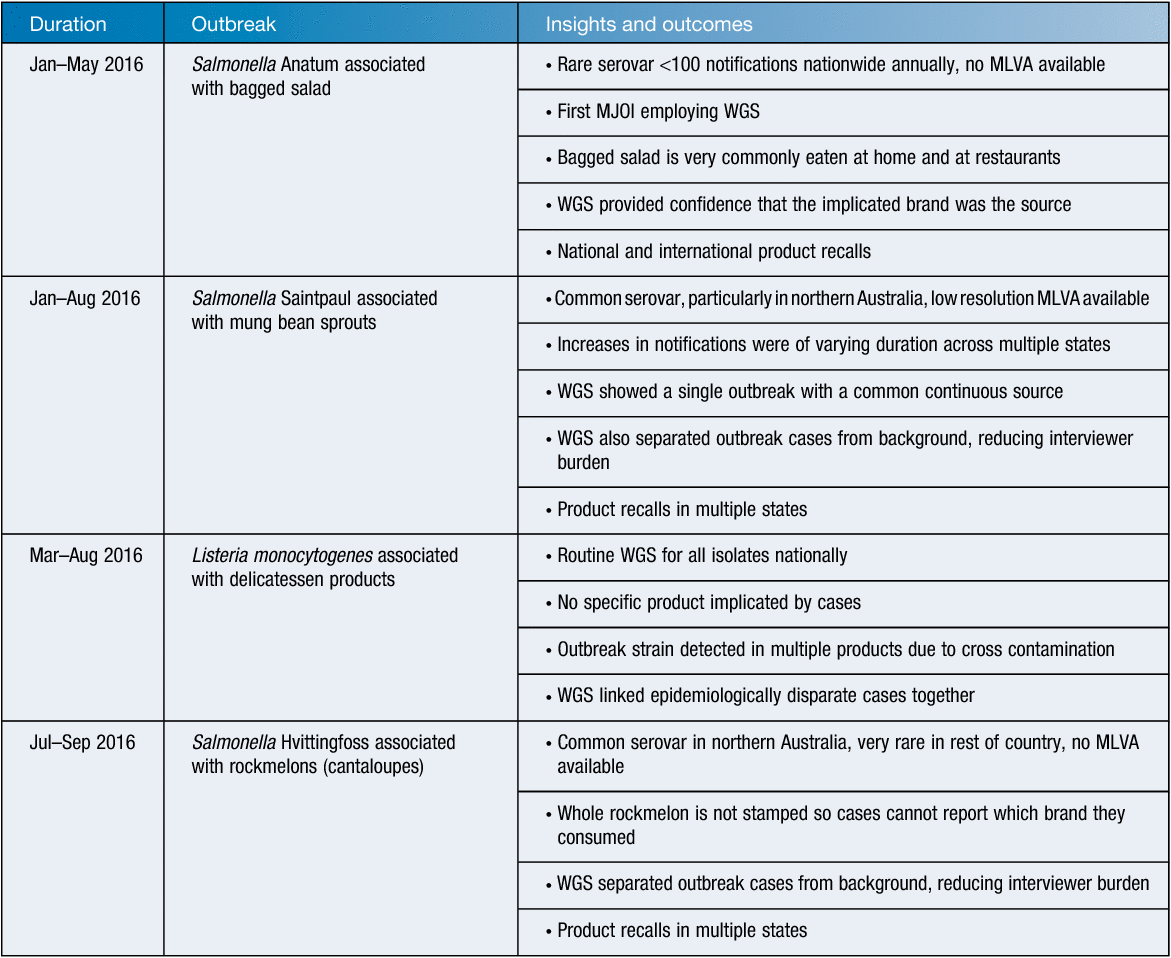
In 2016, OzFoodNet conducted four complex MJOIs that included the aforementioned listeriosis MJOI, and three salmonellosis outbreaks associated with fresh produce. There were over 800 reported human infections as a result of these outbreaks. Three MJOIs were being conducted concurrently between the months of February to May and June to August, testing the resilience of the network. WGS was very valuable in precisely defining the outbreak cases and separating them from background cases, which has been difficult for less prevalent serovars in the past. The WGS definitively linked cases to a source food that was essential for foods that may be frequently eaten but cases commonly are unable to recall specific brands, such as bagged salad, or foods used as a garnish that cases may not remember eating at all, like mung bean sprouts (Table 1).
|
Continuing process improvement is written into the Guidelines, and a structured audit6 is held after every MJOI. An independent facilitator leads a discussion of successes and challenges, and recommendations for improvement are developed. The findings of these audits are fed back into the process and the Guidelines are revised. The latest iteration of the Guidelines incorporating the findings from the 2016 MJOIs was recently endorsed by the Australian Health Protection Principal Committee (AHPPC).
Conclusions
If CIDT increases at the expense of culture methods, OzFoodNet may potentially lose the ability to detect clusters and outbreaks, and community based outbreaks may not be detected at all. The value of reflex culture in light of CIDT is enormous in terms of public health surveillance and for the OzFoodNet network to continue to do the work that it has done for over 15 years. The benefits of the integration of WGS into outbreak investigations have been demonstrated in recent times and this technology will likely continue to be utilised in the investigations of outbreaks of foodborne illness.
Acknowledgements
We thank the Australian Government Department of Health for continued funding and support of OzFoodNet. We thank the many epidemiologists, Masters of Philosophy in Applied Epidemiology scholars, Health graduate trainees, project officers, interviewers and research assistants at each of the OzFoodNet sites. We acknowledge the work of various public health professionals and laboratory staff around Australia who interviewed patients, tested specimens, typed isolates and investigated outbreaks. We would particularly like to acknowledge the PHLN expert advisory group on whole genome sequencing and the Australasian Communicable Diseases Genomics Network for establishing national capacity for WGS in public health microbiology; and especially MDU, the Institute of Clinical Pathology and Medical Research and Queensland Health Scientific Services, for their help and guidance with foodborne disease surveillance and communicable diseases genomics. The quality of their work remains the foundation of OzFoodNet.
Further information on the network including the Guidelines for the epidemiological investigation of multi-jurisdictional outbreaks that are potentially foodborne is available from the OzFoodNet website: http://health.gov.au/internet/main/publishing.nsf/Content/cdna-ozfoodnet.htm.
References
[1] Green, M. and Fitzsimmons, G. (2013) The OzFoodNet story: 2000 to present day. Microbiol. Aust. 34, 59–62.| The OzFoodNet story: 2000 to present day.Crossref | GoogleScholarGoogle Scholar |
[2] Kwong, J.C. et al. (2016) Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 54, 333–342.
| Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xps1ensLs%3D&md5=eedd228d2a7d589510de704de34b1417CAS |
[3] Sloan-Gardner, T. (2014) Applied epidemiology of infectious diseases in Australia, Master of Philosophy in Applied Epidemiology (MAE) thesis. https://openresearch-repository.anu.edu.au/bitstream/1885/110689/2/b37326831-Sloan-Gardner_T.pdf
[4] Communicable Diseases Branch, Health Protection NSW (2016) NSW OzFoodNet Annual Surveillance Report.
[5] Kwong, J.C. et al. (2016) Sharing is caring: international sharing of data enhances genomic surveillance of Listeria monocytogenes. Clin. Infect. Dis. 63, 846–848.
| Sharing is caring: international sharing of data enhances genomic surveillance of Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar |
[6] Dalton, C.B. et al. (2009) A structured framework for improving outbreak investigation audits. BMC Public Health 9, 472.
| A structured framework for improving outbreak investigation audits.Crossref | GoogleScholarGoogle Scholar |
Biographies
Michelle Harlock is currently the OzFoodNet Epidemiologist in Tasmania where she has worked for the past five years. Prior to this she was the OzFoodNet Epidemiologist in the Northern Territory and also spent nearly 10 years as a microbiology scientist in clinical pathology laboratories. She enjoys her work, but also spending time with her children and exploring her beautiful adopted home state of Tasmania.
Anthony Draper has been the OzFoodNet Epidemiologist in the Northern Territory since 2012. Prior to that, he was a medical scientist in Timor-Leste, Solomon Islands, the Northern Territory and Queensland.
Ben Polkinghorne is the OzFoodNet Coordinating Epidemiologist in the Australian Government Department of Health. He has worked in this role since mid-2014. Prior to that he was the NSW OzFoodNet Epidemiologist for 12 months.
Robyn Leader has worked in OzFoodNet Central since 2004, including on projects to estimate the burden of foodborne disease and to establish genomic surveillance in Australia. Prior to that, she worked on antimicrobial resistance in the Australian Government Department of Health.