Virus discovery in bats
Rebecca I Johnson A B and Ina L Smith A CA CSIRO Australian Animal Health Laboratory, 5 Portarlington Road, East Geelong, Vic. 3219, Australia
B Email: Rebecca.Johnson@csiro.au
C Email: Ina.Smith@csiro.au
Microbiology Australia 38(1) 25-27 https://doi.org/10.1071/MA17008
Published: 9 February 2017
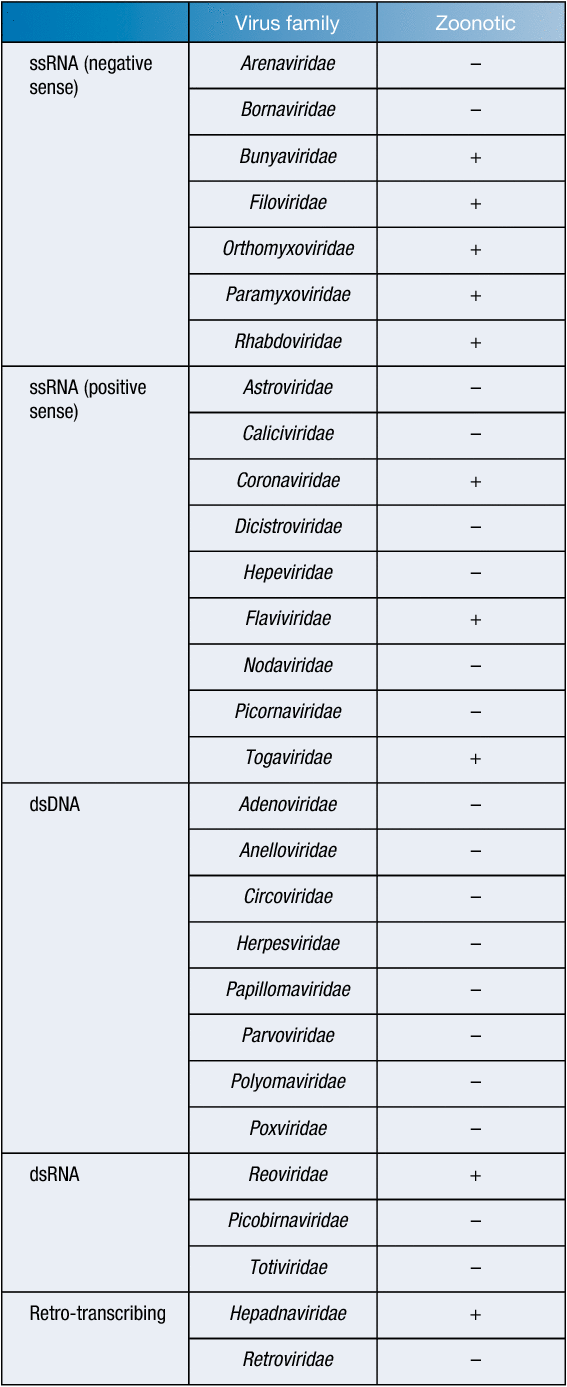
Comprising approximately 20% of known mammalian species, bats are abundant throughout the world1. In recent years, bats have been shown to be the reservoir host for many highly pathogenic viruses, leading to increased attempts to identify other zoonotic bat-borne viruses. These efforts have led to the discovery of over 200 viruses in bats and many more viral nucleic acid sequences from 27 different viral families2,3 (Table 1). Over half of the world’s recently emerged infectious diseases originated in wildlife15, with the genetic diversity of viruses greater in bats than in any other animal16. As humans continue to encroach on the habitat of bats, the risk of spillover of potentially zoonotic viruses is also continuing to increase. Therefore, the surveillance of bats and discovery of novel pathogens is necessary to prepare for these spillover events17.
|
Not only does virus discovery increase our understanding of the role that bats play in emerging infectious diseases, it also allows the development of diagnostic tools resulting in a much more efficient response if a spillover event occurred, reducing both the economic and public health impact of the virus. Virus discovery is important for identifying potential zoonotic threats and can assist with the characterisation of already emerged zoonotic viruses, as well as providing phylogenetic evidence for the origin and evolution of these viruses; for example the potential bat origin of primate hepadnaviruses5.
Advancements in technology have also contributed to the increased rate of virus discovery, with molecular techniques now overtaking serological methods and virus isolation18. Improvement in the accessibility of next generation sequencing has allowed the development of unbiased methods of analysing bat specimens as well as more rapid characterisation of novel viruses. However, next generation sequencing is not suitable for all experimental aims, such as when the targeted discovery of particular viral families is required19.
The bat sampling method can affect which viruses are able to be detected and can result in a bias towards particular families of viruses. The bat specimen used for discovery is an important consideration, as well as the time of year these specimens are collected, the intervals between collections, the species of bat to be targeted and the ecology of the bat species, especially as not all viruses are continually shed in the population. In the case of Marburg virus, peaks of shedding were seen during birthing seasons as these months coincided with a peak in infection in 6-month-old juvenile bats20.
Although lethal sampling of bats may be necessary for virus discovery from particular viral families, non-lethal sampling has resulted in the discovery of a greater number of novel viruses across a similar number of studies18. Bat urine and faeces have been favoured as non-invasive samples for virus discovery, however active bat catching and sampling can give more accurate calculations of viral prevalence. In the case of Hendra virus, urine was the most significant form of virus transmission, with higher titres of virus seen in urine compared to specimens such as nasal swabs, faecal samples and serum21. Pooled urine can be collected from plastic sheets laid below bat colonies and stored in a viral transport medium at −80°C for later analysis22. These samples can then be analysed in multiple different ways depending on the chosen method for virus discovery.
Molecular techniques such as pan-viral family PCR are useful for targeted discovery of viruses. This involves amplifying a region of the genome that is highly conserved across that viral family using degenerate primers23,24. In one study, this approach was employed to detect sequences of 66 new viruses from the Paramyxoviridae family from bats and rodents around the world25. In this example, pan-viral family PCR detected paramyxovirus sequences, including in bats that yielded no positive results when their pooled serum samples were analysed by next generation sequencing. However, it is possible that the negative results by next generation sequencing were due to low concentrations of virus in the blood rather than significantly lower sensitivity25. The primers utilised by pan-viral family PCR can only detect viruses that are related to previously identified viruses. In an attempt to reduce the bias introduced by sequence specific primers, multiple different primer sets and methods can be utilised for the same samples25. Although this approach has led to the discovery of many novel viruses, other methods provide a hypothesis-free approach.
Next generation sequencing has become increasingly more accessible as a method for virus discovery, although it is still more expensive than other molecular methods and requires bioinformatics knowledge to correctly analyse the raw data and generate a consensus genome19. When correctly designed, metagenomic analysis of bat specimens can allow the hypothesis free discovery of many novel viruses, including those that are significantly divergent from previously identified viruses. The high throughput technique also allows efficient screening of a large number of bat specimens. This method was used to identify highly divergent novel rotaviruses in bats in Cameroon that were unlikely to have been successfully detected using the currently available primer combinations26. The sensitivity of high throughput sequencing is continuing to improve for virus discovery, employing techniques such as positive enrichment of samples for virus sequences using probes that cover the genomes of all the viral taxa known to infect vertebrates27. However, this enrichment may reduce the likelihood of discovering novel viruses.
Virus isolation, supported by other molecular detection techniques, continues to play a significant role in the discovery of novel viruses as it allows further characterisation and comparison with other viruses. Virus isolation followed by pan-family PCR was successfully used for the surveillance of Australian pteropid bats and resulted in the discovery of multiple novel paramyxoviruses22. However, not all viruses cause obvious cytopathic effect in cell culture, making it difficult to detect virus growth in cells. Furthermore, the viruses may require very specific cell lines and conditions for growth, if they can even be cultured at all. Bat derived influenza viruses have been detected in Sturnira lilium in Guatemala by pan-influenza virus RT-PCR28, but subsequent attempts at culturing were challenging, due in part to their divergent surface proteins and unique basolateral cell entry mechanism29,30. In vivo isolation methods may also be used, such as the use of suckling mice or knockout mice31.
Virus discovery from bats increases our database of known viruses and is necessary for preparing a rapid response to emerging infectious diseases17. For example, the isolation and characterisation of Hendra virus in 1994 enabled the development of diagnostic assays that played an important role in the identification of Nipah virus during an outbreak of encephalitic disease five years later. Cross-reactivity with antibodies to Hendra virus was observed during initial screening against the unknown virus causing fatal disease in pigs and humans. Then, primers developed against Hendra virus assisted in determining the sequence of Nipah virus32. Virus discovery can also facilitate the development of diagnostic tools and further research into pathogenic determinants of other viruses. It is estimated that each bat species would have to be sampled 7000 times before the viral diversity limit is reached33, so with approximately 1200 species of bat around the world, the discovery of novel viruses in bats has a long way to go.
References
[1] Churchill, S. (1998) Australian Bats. Reed New Holland, Sydney, Australia.[2] Moratelli, R. and Calisher, C.H. (2015) Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz 110, 1–22.
| Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses?Crossref | GoogleScholarGoogle Scholar |
[3] Chen, L. et al. (2014) DBatVir: the database of bat-associated viruses. Database: the journal of biological databases and curation 2014, bau021.
| DBatVir: the database of bat-associated viruses.Crossref | GoogleScholarGoogle Scholar |
[4] Luis, A.D. et al. (2013) A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 280, 20122753.
| A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special?Crossref | GoogleScholarGoogle Scholar |
[5] Drexler, J.F. et al. (2013) Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. USA 110, 16151–16156.
| Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs1SqtbfN&md5=91b19c6a7ebe612dacf1d28ae345f60eCAS |
[6] Dacheux, L. et al. (2014) A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses. PLoS One 9, e87194.
| A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses.Crossref | GoogleScholarGoogle Scholar |
[7] Cogswell-Hawkinson, A. et al. (2012) Tacaribe virus causes fatal infection of an ostensible reservoir host, the Jamaican fruit bat. J. Virol. 86, 5791–5799.
| Tacaribe virus causes fatal infection of an ostensible reservoir host, the Jamaican fruit bat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmslOit7c%3D&md5=84d6ba15a44a6c9cefd8c417645c298dCAS |
[8] Tse, H. et al. (2012) Discovery and genomic characterization of a novel bat sapovirus with unusual genomic features and phylogenetic position. PLoS One 7, e34987.
| Discovery and genomic characterization of a novel bat sapovirus with unusual genomic features and phylogenetic position.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xmt1eht7Y%3D&md5=9547dfc95e8faf43a4e85b8034527323CAS |
[9] Drexler, J.F. et al. (2012) Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J. Virol. 86, 9134–9147.
| Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1CgsLjE&md5=c18d4cdf4ea493efbcd2ac651dd7cc21CAS |
[10] Baker, K.S. and Murcia, P.R. (2014) Poxviruses in bats ... so what? Viruses 6, 1564–1577.
| Poxviruses in bats ... so what?Crossref | GoogleScholarGoogle Scholar |
[11] Lima, F.E. et al. (2015) Genomic characterization of novel circular ssDNA viruses from insectivorous bats in Southern Brazil. PLoS One 10, e0118070.
| Genomic characterization of novel circular ssDNA viruses from insectivorous bats in Southern Brazil.Crossref | GoogleScholarGoogle Scholar |
[12] Canuti, M. et al. (2011) Two novel parvoviruses in frugivorous New and Old World bats. PLoS One 6, e29140.
| Two novel parvoviruses in frugivorous New and Old World bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xms1CjsA%3D%3D&md5=76924cbd8b16ba9dbaa2a940e76bd72dCAS |
[13] Cibulski, S.P. et al. (2014) A novel anelloviridae species detected in Tadarida brasiliensis bats: first sequence of a Chiropteran Anellovirus. Genome Announc. 2, e01028-14.
| A novel anelloviridae species detected in Tadarida brasiliensis bats: first sequence of a Chiropteran Anellovirus.Crossref | GoogleScholarGoogle Scholar |
[14] Yang, X. et al. (2012) A novel totivirus-like virus isolated from bat guano. Arch. Virol. 157, 1093–1099.
| A novel totivirus-like virus isolated from bat guano.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XnvVSrsrs%3D&md5=af3ba1333deb352f4a411dc252140986CAS |
[15] Jones, K.E. et al. (2008) Global trends in emerging infectious diseases. Nature 451, 990–993.
| Global trends in emerging infectious diseases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXit1ygurg%3D&md5=edba0bf575941674db0ed8de6bf6b78bCAS |
[16] Li, Y. et al. (2010) Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 84, 3889–3897.
| Host range, prevalence, and genetic diversity of adenoviruses in bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXkvVyqs7Y%3D&md5=89529d9cd016f249a5a9b39981c53245CAS |
[17] Daszak, P. et al. (2000) Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science 287, 443–449.
| Emerging infectious diseases of wildlife--threats to biodiversity and human health.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXntl2jtw%3D%3D&md5=e700279fe7b87a1d0f0ecf6bcd793171CAS |
[18] Young, C.C. and Olival, K.J. (2016) Optimizing viral discovery in bats. PLoS One 11, e0149237.
| Optimizing viral discovery in bats.Crossref | GoogleScholarGoogle Scholar |
[19] Radford, A.D. et al. (2012) Application of next-generation sequencing technologies in virology. J. Gen. Virol. 93, 1853–1868.
| Application of next-generation sequencing technologies in virology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1Wgs7nP&md5=63c7a8069658264538a047c6d78200caCAS |
[20] Amman, B.R. et al. (2012) Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 8, e1002877.
| Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection.Crossref | GoogleScholarGoogle Scholar |
[21] Edson, D. et al. (2015) Routes of Hendra virus excretion in naturally-infected flying-foxes: implications for viral transmission and spillover risk. PLoS One 10, e0140670.
| Routes of Hendra virus excretion in naturally-infected flying-foxes: implications for viral transmission and spillover risk.Crossref | GoogleScholarGoogle Scholar |
[22] Barr, J. et al. (2015) Isolation of multiple novel paramyxoviruses from pteropid bat urine. J. Gen. Virol. 96, 24–29.
| Isolation of multiple novel paramyxoviruses from pteropid bat urine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtVGns7fM&md5=b4e3c602bf49dd1c8e1143151172e856CAS |
[23] Tong, S. et al. (2008) Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J. Clin. Microbiol. 46, 2652–2658.
| Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVyms73M&md5=69bf79bf60ef1d7d7d74859101cc76b8CAS |
[24] Rose, T.M. (2005) CODEHOP-mediated PCR – a powerful technique for the identification and characterization of viral genomes. Virol. J. 2, 20.
| CODEHOP-mediated PCR – a powerful technique for the identification and characterization of viral genomes.Crossref | GoogleScholarGoogle Scholar |
[25] Drexler, J.F. et al. (2012) Bats host major mammalian paramyxoviruses. Nat. Commun. 3, 796.
| Bats host major mammalian paramyxoviruses.Crossref | GoogleScholarGoogle Scholar |
[26] Yinda, C.K. et al. (2016) Novel highly divergent reassortant bat rotaviruses in Cameroon, without evidence of zoonosis. Sci. Rep. 6, 34209.
| Novel highly divergent reassortant bat rotaviruses in Cameroon, without evidence of zoonosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsFyntbfP&md5=9667eb7a0124193a4f048db467111e42CAS |
[27] Briese, T. et al. (2015) Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio 6, e01491–15.
| Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis.Crossref | GoogleScholarGoogle Scholar |
[28] Tong, S. et al. (2012) A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 109, 4269–4274.
| A distinct lineage of influenza A virus from bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XkslCksL0%3D&md5=0c04e3d04b54011533aabd47b203b541CAS |
[29] Sun, X. et al. (2013) Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Reports 3, 769–778.
| Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjtVShsLc%3D&md5=08b1def84a8e440acb115bd71a95cb21CAS |
[30] Moreira, E.A. et al. (2016) Synthetically derived bat influenza A-like viruses reveal a cell type- but not species-specific tropism. Proceedings of the National Academy of Sciences of the United States of America.
[31] Lipkin, W.I. (2010) Microbe hunting. Microbiology and molecular biology reviews Microbiol. Mol. Biol. Rev. 74, 363–377.
| Microbe hunting. Microbiology and molecular biology reviewsCrossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsVSqurnM&md5=26b0796bcd1fe19fc52f3e0cf6168d6dCAS |
[32] Harcourt, B.H. et al. (2000) Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271, 334–349.
| Molecular characterization of Nipah virus, a newly emergent paramyxovirus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXktVKqtL8%3D&md5=4eeccdeb261e0202d84d21f5ea212d40CAS |
[33] Anthony, S.J. et al. (2013) A strategy to estimate unknown viral diversity in mammals. MBio 4, e00598–13.
| A strategy to estimate unknown viral diversity in mammals.Crossref | GoogleScholarGoogle Scholar |
Biographies
Rebecca I Johnson is a PhD student at the CSIRO Australian Animal Health Laboratory. She has an interest in emerging diseases and her work focuses on the characterisation of novel viruses isolated from pteropid bats.
Ina L Smith is a senior research scientist at the CSIRO Australian Animal Health Laboratory. With a strong background in traditional and molecular virology, her research has focused on viral discovery and characterisation, primarily from bats but also from mammals and birds.


