Anaerobic microorganisms and bioremediation of organohalide pollution
Matthew Lee A , Chris Marquis A , Bat-Erdene Judger A and Mike Manefield A BA School of Biotechnology and Biomolecular Sciences
University of New South Wales
Sydney, NSW 2052, Australia
Tel: +61 2 9385 1780
B Email: manefield@unsw.edu.au
Microbiology Australia 36(3) 125-128 https://doi.org/10.1071/MA15044
Published: 11 August 2015
Organohalide pollution of subsurface environments is ubiquitous across all industrialised countries. Fortunately, strictly anaerobic microorganisms exist that have evolved using naturally occurring organohalides as their terminal electron acceptor. These unusual organisms are now being utilised to clean anthropogenic organohalide pollution.
Subsurface water systems are a precious resource particularly in Australia where they are used for agricultural, industrial and domestic purposes including for drinking. However, a large range of organic and inorganic pollutants often compromise this precious resource, such as nitrates from overuse of fertilisers, heavy metals from the mining industry and BTEX compounds from petroleum spillages.
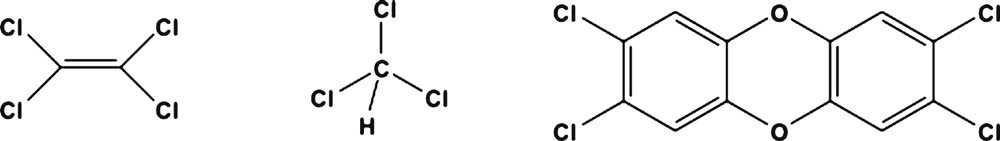
Bioremediation of organohalides has garnered a significant amount of research interest over the past 20 years1. Organohalides are a large family of compounds ranging from simple C1 and C2 aliphatics to complicated polyaromatic structures such as dioxins (Figure 1). The chemical and physical characteristics of these compounds have enabled them to be used in a large range of applications. Some of the more notable examples include: vinyl chloride (VC) in the manufacture of polyvinyl chloride (PVC), perchloroethene (PCE) as dry cleaning solvent and trichloromethane (chloroform; CF) as precursor chemical in the production of chlorofluorocarbon refrigerant gases. Because of their utility, these compounds have been synthesised in enormous quantities. For example PCE production in the USA peaked at 320 million kg per annum in the 1980s2. Industrial activity on such a large scale has resulted in the discharge of organohalides into the environment, where because of their high density and limited water solubility, tend to reside as solvent pools within subsurface water systems. These subsurface solvent pools are known as dense non-aqueous phase liquid (DNAPL) source zones. The DNAPL slowly dissolves into the surrounding water creating a toxic solvent plume radiating out from the source zone in the direction of the groundwater flow. To give some perspective on the extent of organohalide pollution, of the 1319 priority contaminated sites listed by the USA-EPA, 819 (61%) of them are polluted with PCE3.
|
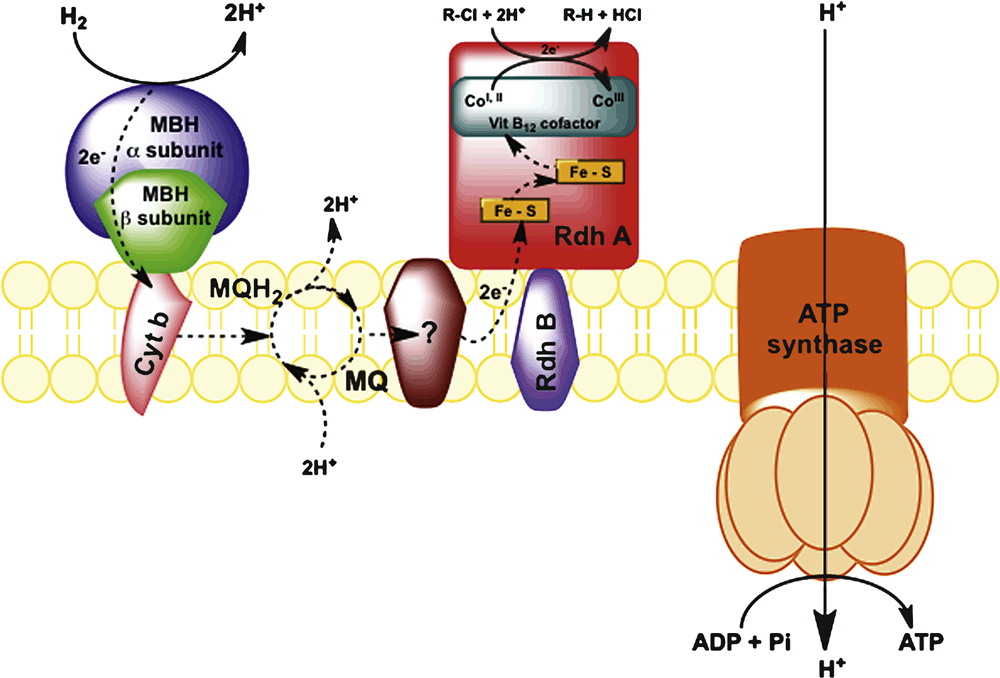
Fortunately, strictly anaerobic bacteria exist that have evolved to utilise organohalides as terminal electron acceptors. These organisms predate anthropogenic organohalide production and owe their existence to a wide range of naturally occurring organohalides in the environment4. Their metabolism results in the elimination of a halogen atom, which ultimately yields a much less toxic hydrocarbon. Organohalide respiring bacteria (ORB) perform the dehalogenation reaction by virtue of enzymes known as reductive dehalogenases that are constructed around a cobalt-containing cobalamin cofactor at the active site. The cobalt atom in its reduced form is a strong nucleophile that attacks the carbon atom in a carbon-halogen bond. In most ORB, the electrons that are ultimately transferred to the organohalide are acquired from the oxidation of hydrogen. The electrons are transferred via a series of membrane associated quinones and cytochrome-like proteins to the reductive dehalogenase (RdhA). The transfer of electrons generates a proton motive force and subsequently ATP5 (Figure 2).
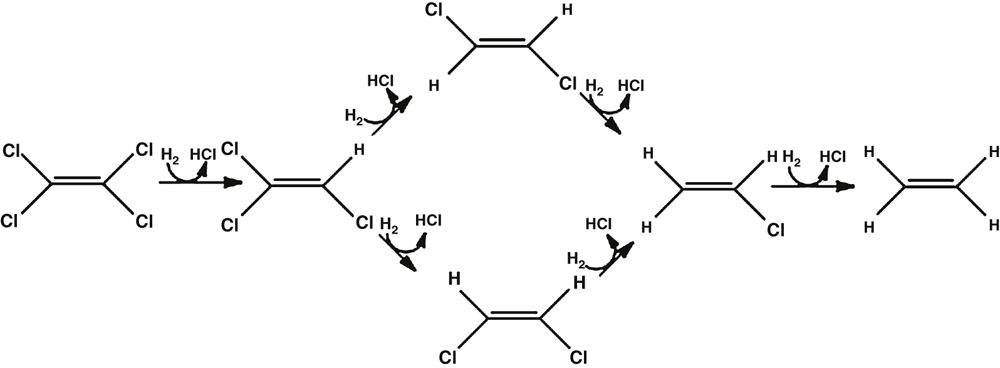
The first ORB reported about 30 years ago transformed 3-chlorobenzoic acid to benzoic acid6. However, the greatest advance in terms of organohalide bioremediation was the discovery of PCE-dechlorinating mixed cultures in the mid 1980s. These cultures produced various daughter compounds ranging from cis-dichloroethene and VC to ethane7,8. Isolation and characterisation of organisms capable of utilising PCE and trichloroethene (TCE) as terminal electron acceptors revealed the mechanism behind the dechlorination reactions observed in the mixed cultures. Strains from genera including Dehalospirillium, Desulfitobacterium, Desulfuromonas and Dehalobacter were shown to be capable of partial dechlorination of PCE and TCE to cis-DCE and the highly toxic carcinogen VC9–13 (Figure 3).
For bioremediation, the production of partially chlorinated toxic daughter compounds is an undesirable outcome. Therefore, the discovery of Dehalococcoides mccartyii strain 19514, which could transform vinyl chloride to ethene, was extremely important, as this made bioremediation of chlorinated ethanes and ethenes a viable option. Dehalococcoides strains since that time have become synonymous with bioremediation because of their versatility. Dehalococcoides strains harbor up to 32 reductive dehalogenase homologs that are capable of dehalogenating not only halogenated C2 aliphatics, but also halogenated aromatic compounds such as hexachlorobenzene and chlorinated dioxins15–17.
Dehalococcoides resides in the phylum Chloroflexi, to which the class Dehalococcoidetes was added to accommodate organohalide respiring members18. Dehalococcoides cells morphologically are unusual being disc shaped and only 0.3–1 µm in diameter and 0.2 µm thick. Another unusual feature of Dehalococcoides is the apparent lack of peptidoglycan layer in the cell-wall a feature typical of most bacteria18. Electron micrographs have revealed an unusual cell-wall S-layer resembling that found in archaea19. Dehalococcoides like most ORB and other strict anaerobes operate only at very low redox potentials (<110 mV)18. Therefore, minute traces of oxygen quickly curtail dehalogenating activity. This presents a difficulty in cultivating these types of organisms under laboratory conditions, where equipment and chemicals must be scrupulously deoxygenated in anaerobic chambers or with chemical reducing agents before they can be used.
Over the past 20 years numerous obligate and facultative ORB have been reported, and between them, these bacteria can use the vast majority of anthropogenic chlorinated ethanes and ethenes as sole electron acceptors (reviewed in Koenig et al.3). However, in reality, many organohalide-polluted sites contain more than one halogenated compound, which poses a challenge for ORB. The presence of chlorinated methanes in the solvent plume has been problematic, as they are strong inhibitors of most anaerobic microbial processes including organohalide respiration20,21. CF is particularly problematic as it is a widespread pollutant and highly recalcitrant, with a half-life of 3500 years in groundwater in the absence of appropriate microbes22. Until recently, the presence of CF has required site pretreatment for its removal, or the employment of abiotic strategies such as “pump and treat” or the installation of permeable reactive iron barriers23. However, recent discoveries of CF respiring Dehalobacter and Desulfitobacterium strains that transform CF to dichloromethane (DCM) have opened many CF-impacted sites to bioremediation strategies24–26. Although DCM is not a favourable intermediate, recently Dehalobacter strains have been identified that can use DCM as a sole source of organic carbon and electrons, producing acetate, hydrogen and carbon dioxide. Both the CF respiring and DCM fermenting Dehalobacter strains have been shown to work together to effect complete dechlorination of CF26,27.
In conclusion, in subsurface environments where oxygen is in limited supply, microorganisms have evolved to use a multitude of alternative electrons acceptors for energy conservation. Organohalides are naturally occurring molecules in which the carbon halide bond is in a high oxidation state, hence bacteria have evolved to use these compounds as terminal electron acceptors in a process called organohalide respiration. These organisms have been exploited to remediate sub-surface environments that have been polluted with large quantities of anthropogenic organohalides. Research over the past 20 years has revealed numerous genera that are capable of organohalide respiration, with a large array of reductive dehalogenases that can tackle most organohalides found at polluted sites. However, knowledge gaps still exist around how ORB interact with other cohabiting microorganisms such as those supplying hydrogen and cobalamin cofactors.
References
[1] Lovley, D.R. (2003) Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1, 35–44.| Cleaning up with genomics: applying molecular biology to bioremediation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXptFWlu7w%3D&md5=1caab38bf2eabc199062585f093f810eCAS | 15040178PubMed |
[2] Doherty, R.E. (2000) A history of the production and use of carbon tetrachloride, tetrachloroethylene, trichloroethylene and 1, 1, 1-trichloroethane in the United States: part 2--trichloroethylene and 1, 1, 1-trichloroethane. Environ. Forensics 1, 83–93.
| A history of the production and use of carbon tetrachloride, tetrachloroethylene, trichloroethylene and 1, 1, 1-trichloroethane in the United States: part 2--trichloroethylene and 1, 1, 1-trichloroethane.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xis1agur0%3D&md5=4d2805aa7f5f24447be32749b8055cf7CAS |
[3] Koenig, J. et al. (2015) Aliphatic organochlorine degradation in subsurface environments. Rev. Environ. Sci. Biotechnol. 14, 49–71.
| Aliphatic organochlorine degradation in subsurface environments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtlKht7nJ&md5=4356356b7a265e22dd6ab1f7334d7f5aCAS |
[4] Gribble, G.W. (1992) Naturally occurring organohalogen compounds--a survey. J. Nat. Prod. 55, 1353–1395.
| Naturally occurring organohalogen compounds--a survey.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38XmtVOhtbc%3D&md5=6633f577b44a8be1ccadfa3710777695CAS |
[5] Mohn, W.W. and Tiedje, J.M. (1992) Microbial reductive dehalogenation. Microbiol. Rev. 56, 482–507.
| 1:CAS:528:DyaK3sXjsFCkuw%3D%3D&md5=09f313e0459914126a00ca3b000cab47CAS | 1406492PubMed |
[6] DeWeerd, K.A. et al. (1990) Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium. Arch. Microbiol. 154, 23–30.
| Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3cXlt12itL0%3D&md5=b6437bf40e26c57d166f1cd434db8c8eCAS |
[7] Vogel, T.M. and McCarty, P.L. (1985) Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl. Environ. Microbiol. 49, 1080–1083.
| 1:CAS:528:DyaL2MXkt1Kqs7Y%3D&md5=2d50ac2c14ef08996995674c572ec894CAS | 3923927PubMed |
[8] DiStefano, T.D. et al. (1992) Hydrogen as an electron donor for dechlorination of tetrachloroethene by an anaerobic mixed culture. Appl. Environ. Microbiol. 58, 3622–3629.
| 1:CAS:528:DyaK3sXlsFyjug%3D%3D&md5=e98aca7c4082b24889e7ba5c4f2ff5f5CAS | 1482184PubMed |
[9] Scholz-Muramatsu, H. et al. (1995) Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163, 48–56.
| Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXkvVOhsL0%3D&md5=79787d130d8874163151a291df99fce9CAS |
[10] Gerritse, J. et al. (1996) Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165, 132–140.
| Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XisVKqtrg%3D&md5=58ff1c84b2996735096a0ce7b6976b2aCAS | 8593100PubMed |
[11] Krumholz, L.R. (1997) Desulfuromonas chloroethenica sp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors. Int. J. Syst. Bacteriol. 47, 1262–1263.
| Desulfuromonas chloroethenica sp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXntV2qu7s%3D&md5=e7ae3c947d85d1052f9096ec63cc2df4CAS |
[12] Miller, E. et al. (1997) Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 168, 513–519.
| Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXnvFWitbs%3D&md5=24d344fd5a32dc8add27b6a5220bcb7eCAS | 9385143PubMed |
[13] Holliger, C. et al. (1998) Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra-and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169, 313–321.
| Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra-and trichloroethene in an anaerobic respiration.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXivVWgtbs%3D&md5=232cfe5e13ca5f368071be075088d1faCAS | 9531632PubMed |
[14] Maymó-Gatell, X. et al. (2001) Reductive dechlorination of cis-1, 2-dichloroethene and vinyl chloride by ‘Dehalococcoides ethenogenes’ . Environ. Sci. Technol. 35, 516–521.
| Reductive dechlorination of cis-1, 2-dichloroethene and vinyl chloride by ‘Dehalococcoides ethenogenes’ .Crossref | GoogleScholarGoogle Scholar | 11351722PubMed |
[15] Adrian, L. et al. (2000) Bacterial dehalorespiration with chlorinated benzenes. Nature 408, 580–583.
| Bacterial dehalorespiration with chlorinated benzenes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXovFWrsLc%3D&md5=0eb3de0cb00523f331ab38834d30409eCAS | 11117744PubMed |
[16] Bunge, M. et al. (2003) Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421, 357–360.
| Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXlsVagug%3D%3D&md5=7408edb532201a4b12a6de31e3d9ffd4CAS | 12540897PubMed |
[17] Kube, M. et al. (2005) Genome sequence of the chlorinated compound–respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273.
| Genome sequence of the chlorinated compound–respiring bacterium Dehalococcoides species strain CBDB1.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVOhu7jK&md5=914bccc3d793106fc8903a785191249aCAS | 16116419PubMed |
[18] Löffler, F.E. et al. (2013) Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635.
| Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi.Crossref | GoogleScholarGoogle Scholar | 22544797PubMed |
[19] Kandler, O. (1993) Cell wall biochemistry and three-domain concept of life. Syst. Appl. Microbiol. 16, 501–509.
| Cell wall biochemistry and three-domain concept of life.Crossref | GoogleScholarGoogle Scholar |
[20] Bagley, D.M. et al. (2000) Acclimation of anaerobic systems to biodegrade tetrachloroethene in the presence of carbon tetrachloride and chloroform. Water Res. 34, 171–178.
| Acclimation of anaerobic systems to biodegrade tetrachloroethene in the presence of carbon tetrachloride and chloroform.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXht1ehsg%3D%3D&md5=8c0d18f2711b59d3f0e89de6243b1d8aCAS |
[21] Futagami, T. et al. (2006) Effects of chloromethanes on growth of and deletion of the pce gene cluster in dehalorespiring Desulfitobacterium hafniense strain Y51. Appl. Environ. Microbiol. 72, 5998–6003.
| Effects of chloromethanes on growth of and deletion of the pce gene cluster in dehalorespiring Desulfitobacterium hafniense strain Y51.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XpvVKjtr8%3D&md5=fa04d463e85d6f206a40767886a4e541CAS | 16957221PubMed |
[22] Mabey, W. and Mill, T. (1978) Critical review of hydrolysis of organic compounds in water under environmental conditions. J. Phys. Chem. Ref. Data 7, 383–398.
| 1:CAS:528:DyaE1cXlsFaitrw%3D&md5=773bbb0dacfec19a8aac60ec6d1d88c7CAS |
[23] Lee, M. et al. (2015) Relative Contributions of Dehalobacter and Zerovalent Iron in the Degradation of Chlorinated Methanes. Environ. Sci. Technol. 49, 4481–4489.
| Relative Contributions of Dehalobacter and Zerovalent Iron in the Degradation of Chlorinated Methanes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXktlSjtL0%3D&md5=3cee68554a5f425051f6e84ea3916ea9CAS | 25764054PubMed |
[24] Ding, C. et al. (2014) A Desulfitobacterium sp. strain PR reductively dechlorinates both 1,1,1-trichloroethane and chloroform. Environ. Microbiol. 16, 3387–3397.
| A Desulfitobacterium sp. strain PR reductively dechlorinates both 1,1,1-trichloroethane and chloroform.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXitVeksLfJ&md5=ec15bb2550575b81fe6289c6f1fdba86CAS | 24428759PubMed |
[25] Grostern, A. et al. (2010) Chloroform respiration to dichloromethane by a Dehalobacter population. Environ. Microbiol. 12, 1053–1060.
| Chloroform respiration to dichloromethane by a Dehalobacter population.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmtFCrsL4%3D&md5=b6215ecfd7fcbc40c62cb8562841465fCAS | 20089043PubMed |
[26] Lee, M. et al. (2012) Complete chloroform dechlorination by organochlorine respiration and fermentation. Environ. Microbiol. 14, 883–894.
| Complete chloroform dechlorination by organochlorine respiration and fermentation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmvFGgsLw%3D&md5=b2c75ae2c87b0818afcf8913c646293bCAS | 22118646PubMed |
[27] Justicia-Leon, S.D. et al. (2014) Bioaugmentation with distinct Dehalobacter strains achieves chloroform detoxification in microcosms. Environ. Sci. Technol. 48, 1851–1858.
| Bioaugmentation with distinct Dehalobacter strains achieves chloroform detoxification in microcosms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXislGktw%3D%3D&md5=c0e6cce5e0f848397937c6f15ec35006CAS | 24392834PubMed |
Biographies
Matthew Lee is a Senior Research Associate at UNSW. His research interests include microbiology in subsurface environments, particularly chlorinated methane metabolising bacteria.
Chris Marquis is a bioprocess engineer who runs the UNSW Recombinant Products Facility at UNSW and has interests in applications of microbiology and protein.
Bat-Erdene Judger is a PhD student at UNSW. His research project is to heterologous production of functional reductive dehalogenases from Dehalobacter and Dehalococcoides species.
Mike Manefield is an Associate Professor at UNSW. His interests are in environmental microbiology and biotechnology development.