Predicting genome variations between passages of Clostridium difficle by ribotypes
Volker GürtlerSchool of Applied Science
RMIT University
Bundoora, Vic. 3152, Australia
Email: volkergurtler@gmail.com
Microbiology Australia 36(3) 109-110 https://doi.org/10.1071/MA15038
Published: 11 August 2015
Ribotyping is the most widely used method for differentiating strains of Clostridium difficile for epidemiological studies and infection control. Recently there have been calls for standardisation of the technique to which sophisticated technical solutions have been offered. The present note offers a solution for standardisation based on conserved rrn operon Type-specific flanking genes. Furthermore, this technique can be used to detect Type-specific rrn operon deletions in passages from a single strain of C. difficile.
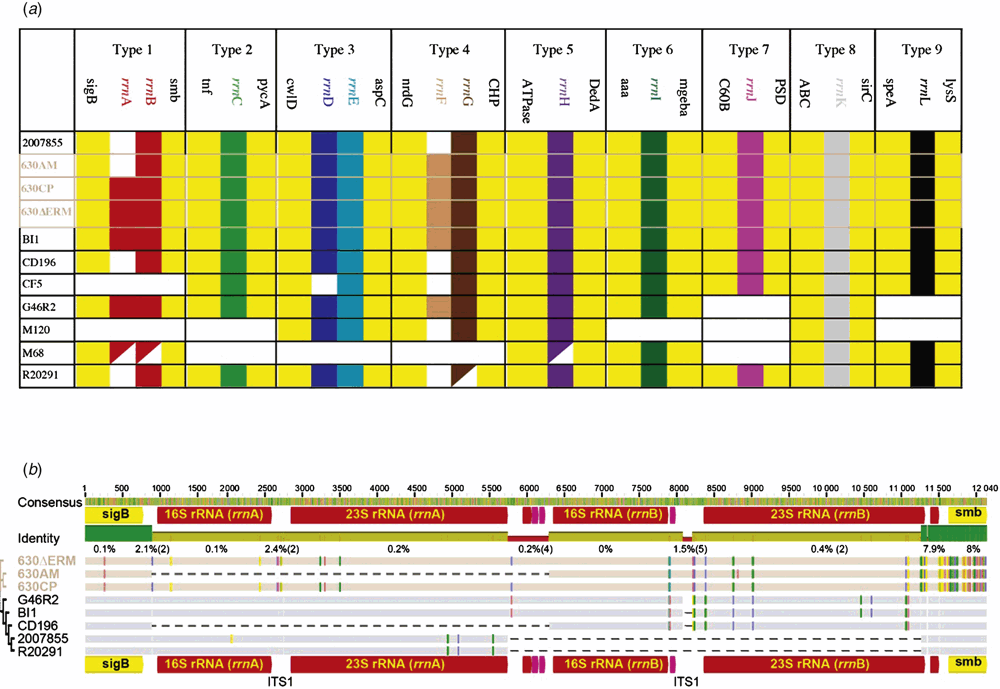
The ribosomal RNA operon is present in up to 12 copies per genome in Clostridium difficile1. Since the early 1990s it has been used for ‘ribotyping’ strains of C. difficile by exploiting sequence variations between operons on the same genome and between operons from different strains2. Detailed analysis of the main ribotypes for which whole genome sequencing (WGS) are available has been performed1 and will not be presented here. Rather, it will be shown how the rrn operons are predictably related to each other between strains by their flanking genes according to Type position on the genome. I will draw on two recent studies3,4 of the well characterised 630 strain that show that according to ribotype, the relatedness of flanking genes (Figure 1) makes it possible to reliably show that rrn operons have been lost and gained between passages of the same strain (e.g. Figure 1a, rrnA is absent in 630AM but present in 630CP and 630ΔERM).
Many ribotyping schemes have been used and reported for C. difficile1 and there is increasing interest in standardisation of methodology with a recent Internationally-Standardised, High-Resolution Capillary Gel-Based Electrophoresis PCR-Ribotyping Protocol providing the technical basis5. However, this protocol does not address the fundamental underlying issue of how the prediction of the systematic similarities and differences in operons can be treated in this way for typing bacterial strains6. The purpose of this short note is to demonstrate how the analysis of rrn Types can be standardised, with simplicity, to provide more reliable typing information, just by including the flanking genes of each rrn operon. By including this information a surprising amount of detail can be obtained regarding the insertion of whole operons between strains7.
First, the genomic position of all the rrn operons can be generalised between strains according to their proximity to Type-specific flanking genes (Figure 1a). This makes it possible to identify specifically deletions, insertions and double rrn operons in some strains of affected Types. The insertions and deletions affect whole genes (16S, 23S and 5S) as well as extragenic regions (ITS1, pre-16S and post-5S). However, all specific Types have identical genes flanking their rrn operons (e.g. Type 1 has sigB and smb in all strains except CF5 and M120). The only exception is when the flanking genes are not present and in this case the rrn operons are absent too. Therefore in these strains the whole Type is missing or rearranged. As can be seen, the genes directly flanking the rrn operons are good for identifying rrn Types and their associated deletions, insertions and rearrangements.
Second, the only Type that has been duplicated in the 630ΔERM strain can be reliably identified and tracked in the related strains as ‘Type 1’ (Figure 1b). Even though the deletion of rrnA (16S and 23S genes) was useful for differentiation of the three CD630 passages it was not specific to CD630, also occurring in CD196 (Figure 1a). But of particular note was the observation that Single Nucleotide Polymorphisms (SNPs) in the two flanking genes (Figure 1b, Open Reading Frames (ORFs) ‘sigB’ and ‘smb’ labelled yellow) could be clearly used to differentiate the three 630 passages from the other five strains (G46R2, BI1, CD196, 2007855 and R20291) with smb having up to 80 times more SNPs than the rrn genes.
This method of analysis has determined that in the rrn operon of ‘Type 1’ of C. difficile strains, the duplication of rrnA has occurred in multiple strains of different lineages (i.e. in strains 630AM and CD196). There is potential for the method outlined here to be used to differentiate between passages of other C. difficile strains and strains of different bacterial species.
References
[1] Gürtler, V. and Grando, D. (2013) New opportunities for improved ribotyping of C. difficile clinical isolates by exploring their genomes. J. Microbiol. Methods 93, 257–272.| New opportunities for improved ribotyping of C. difficile clinical isolates by exploring their genomes.Crossref | GoogleScholarGoogle Scholar | 23545446PubMed |
[2] Gürtler, V. (1993) Typing of Clostridium difficile strains by PCR-amplification of variable length 16S-23S rDNA spacer regions. J. Gen. Microbiol. 139, 3089–3097.
| Typing of Clostridium difficile strains by PCR-amplification of variable length 16S-23S rDNA spacer regions.Crossref | GoogleScholarGoogle Scholar | 7510324PubMed |
[3] Riedel, T. et al. (2015) Genome resequencing of the virulent and multidrug-resistant reference strain Clostridium difficile 630. Genome Announc. 3, e00276-15.
| Genome resequencing of the virulent and multidrug-resistant reference strain Clostridium difficile 630.Crossref | GoogleScholarGoogle Scholar | 25858846PubMed |
[4] van Eijk, E. et al. (2015) Complete genome sequence of the Clostridium difficile laboratory strain 630Δerm reveals differences from strain 630, including translocation of the mobile element CTn5. BMC Genomics 16, 31.
| Complete genome sequence of the Clostridium difficile laboratory strain 630Δerm reveals differences from strain 630, including translocation of the mobile element CTn5.Crossref | GoogleScholarGoogle Scholar | 25636331PubMed |
[5] Fawley, W.N. et al. (2015) Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One 10, e0118150.
| Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 25679978PubMed |
[6] Gürtler, V. et al. (2014) Bacterial typing and identification by genomic analysis of 16S–23S rRNA intergenic transcribed spacer (ITS) sequences. In Methods in Microbiology. (Goodfellow, M., Sutcliffe, I., Chun, J., eds). 41; 253–274.
[7] Gürtler, V. et al. (2015) Typing of Vibrio parahaemolyticus environmental and clinical strains by ribosomal rna operon allele-specific flanking genes. Front. Microbiol. , .
Biography
Dr Gürtler’s research interest has been in developing diagnostic tests primarly using the ribosomal RNA operon for the last 25 years. He has recently developed a web site where more information on the techinques discussed in this note can be obtained (www.ribotyping.net). He is also Editor of the Journal of Microbiological Methods (Elsevier).