Australia in the global picture of the molecular epidemiology of Cryptococcus gattii molecular type VGII
Carolina Firacative A , Kennio Ferreira-Paim A B , Luciana Trilles C , David M Engelthaler D and Wieland Meyer A EA Molecular Mycology Research Laboratory, Centre for Infectious Diseases and Microbiology, Sydney Medical School – Westmead Hospital, Marie Bashir Institute for Infectious Diseases and Biosecurity, The University of Sydney, Westmead Millennium Institute, Sydney, NSW, Australia
B Infectious Disease Department, Triangulo Mineiro Federal University, Uberaba, Minas Gerais, Brazil
C Laboratório de Micologia, Instituto Nacional de Infectologia Evandro Chagas (INI), Fundação Oswaldo Cruz (FIOCRUZ), Rio de Janeiro, Brazil
D Translational Genomics Research Institute, Flagstaff, AZ, USA
E Corresponding author. Tel: +61 2 8627 3430, Fax: +61 2 9891 5317, Email: wieland.meyer@sydney.edu.au
Microbiology Australia 36(2) 67-70 https://doi.org/10.1071/MA15023
Published: 17 March 2015
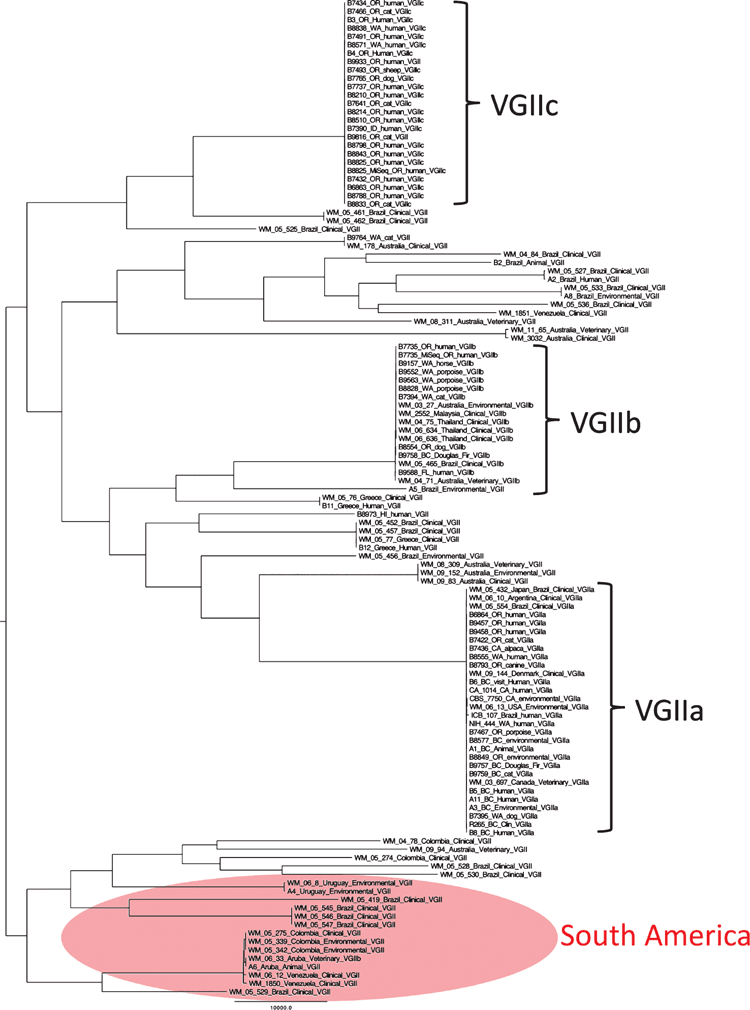
Cryptococcosis, a life-threatening disease of the lung and central nervous system of humans and a broad range of other animals, is caused by the basidiomycetous yeasts Cryptococcus neoformans and C. gattii1. Although most cases of infection in the world are caused by C. neoformans, there is an important prevalence of C. gattii among clinical and veterinary samples in Australia2–4 and the natural habitat of the yeast is strongly associated with native Eucalyptus species4,5, which together position Australia as an endemic area for the less common cryptococcal species. From the numerous C. gattii infections that have been reported in Australia, the molecular type VGII, amongst the four recognised molecular types (VGI-VGIV), has been associated with a high occurrence and a number of small cryptococcosis outbreaks, with most of the isolates belonging to the clonal subtype VGIIb2, which was initially described in 1999 causing part of the ongoing cryptococcosis outbreak on Vancouver Island, British Columbia, Canada6. These findings indicate that Australia is an important stepping-stone in the global dispersion of this outbreak-related subtype and highlight the need for continuous surveillance.
Since it was first reported in Vancouver Island and because of an increased number of human and veterinary C. gattii infections, previously uncommon in North America6,7, C. gattii has been the focus of several studies conducted to elucidate the underlying evolutionary mechanisms involved in its emergence and dispersal in temperate climates and to determine the potential geographical origin of this fungus. At first, two distinct subtypes among the molecular type VGII isolates responsible for the Canadian outbreak were characterised by multilocus sequence typing (MLST), the clinical and environmental common highly virulent VGIIa subtype, and the rarer, less virulent VGIIb subtype8. The early finding that both subtypes have only mating type alpha isolates that were fertile, led to the suggestion that same-sex mating between two alpha cells was the driving force for the emergence of the outbreak8. Within a decade, an additional novel VGII subtype, VGIIc, also with increased virulence, but not associated with the Vancouver Island outbreak, was identified in the Pacific Northwest, which emphasised the continuous emergence of new genotypes among C. gattii7.
MLST analysis9 of a larger number of globally collected clinical, veterinary and environmental isolates showed that the subtypes VGIIa and VGIIb are not only present in North America, but also in South America and Europe, with the subtype VGIIb especially being also found in Asia and Australia2,10,11. Supported by the early association reported between C. gattii and Eucalyptus species4,5 and the extensive introduction of these trees to different places in the world, such as California in the USA, the identification of the subtype VGIIb in Australia, suggested an Australian origin of the outbreak isolates8. However, the presence of VGIIa and VGIIb in South America and the close association of C. gattii with a number of tropical trees that has been reported in this region, indicated strongly that both genotypes originated from South America and have since then been dispersed and introduced on several occasions to other parts of the world, including North America10.
To shed light on the speculations that have arisen on the origins of the outbreak strains, the Molecular Mycology Research Laboratory at the University of Sydney is leading an ongoing research collaboration among different institutions in Australia, South and North America, Asia, Africa and Europe to carry out global epidemiological studies based on MLST typing9. When looking worldwide at the geographical distribution of the genetic diversity, shown by this approach, the highest number of sequence types has been detected so far in South America, while Australia harbors only few sequence types (Figure 1). High levels of genetic diversity are a strong indication of ancestral origins, hence, this extended genetic diversity seems likely to corroborate the idea that the Vancouver Island outbreak subtypes have originated from South America2,10. In contrast, the low genetic diversity combined with the over-representation of some sequence types in Asia and Australia suggest that a clonal expansion has occurred and that the colonisation of such clones may lead to the development of local outbreaks, such as the one observed in a group of sheep in Western Australia4.
|
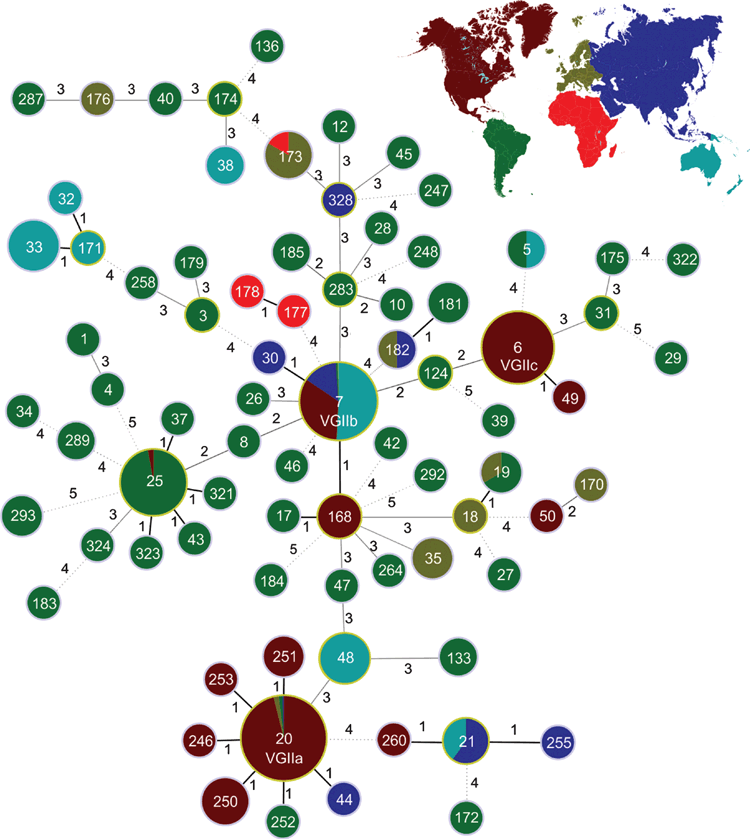
Taking advantage of the more comprehensive view obtained by investigating the whole genome, whole genome sequencing (WGS) was recently performed on 118 C. gattii VGII isolates from five continents, representing most of the MLST haplotypes previously identified2,10–12. Even though the subtypes from the Pacific Northwest of the USA were completely clonal (Figure 2), as shown already by MLST (Figure 1), but with the much greater resolution obtained by WGS, they showed various genetic differences with the other VGII lineages, including mutations, deletions, transpositions, recombination events, and gene content differences which are potentially related to habitat adaptation, virulence, and pathology12. Presence or absence of genes among the different isolates is currently being characterised to understand any functional role, by gene knockout studies carried out in our laboratory. WGS analysis showed again, that i) the highest genetic diversity within the VGII population is present in isolates from South America, ii) the major outbreak genotypes VGIIa and VGIIb are both present in South America, and iii) the Pacific Northwest genotype VGIIc is closely related to a South American isolate, which all support the evolutionary origin and dispersal of C. gattii from this part of the world (Figure 2)12. These findings were independently confirmed in a parallel study investigating 53 different VGII isolates13.
Our current data indicate that Australia is not the origin of the North American outbreaks but a major stepping-stone in the global spread of outbreak-related C. gattii genotypes. As seen with the outbreaks in North America, C. gattii will continue to expand its ecological niche, because there is still a constant and dynamic process driving its evolution. Thus, further characterisation of isolates from regions like Australia, Brazil and Colombia, where a relatively high incidence of cryptococcosis due to C. gattii occurs in some native animals and indigenous human populations3,4,14,15, need to be undertaken in order to better understand the key processes for the emergence of C. gattii epidemics around the globe, especially in regions where the yeast is not thought to be endemic.
References
[1] Kwon-Chung, K.J. et al. (2014) Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 4, a019760.| Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis.Crossref | GoogleScholarGoogle Scholar | 24985132PubMed |
[2] Carriconde, F. et al. (2011) Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population: an emerging outbreak in Australia. PLoS ONE 6, e16936.
| Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population: an emerging outbreak in Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXivFGnuro%3D&md5=978ab520191ba030b5055b275e2f0a8eCAS | 21383989PubMed |
[3] Chen, S. et al. (2000) Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin. Infect. Dis. 31, 499–508.
| Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M%2FksVSlsQ%3D%3D&md5=36adead8a33ccc1d0313a9e375ebc2bcCAS | 10987712PubMed |
[4] Sorrell, T.C. et al. (1996) Natural environmental sources of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 34, 1261–1263.
| 1:STN:280:DyaK283osVCgtA%3D%3D&md5=536e4e83541d33fda4f20c09f900fa55CAS | 8727913PubMed |
[5] Ellis, D.H. and Pfeiffer, T.J. (1990) Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28, 1642–1644.
| 1:STN:280:DyaK3czjvVCmsw%3D%3D&md5=8f2796161036753952400b09efb7baecCAS | 2199524PubMed |
[6] Kidd, S.E. et al. (2004) A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 101, 17258–17263.
| A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtFCrurrJ&md5=db41f89b1b2d860ac2d83c9325a6c504CAS | 15572442PubMed |
[7] Byrnes, E.J. et al. (2010) Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6, e1000850.
| Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States.Crossref | GoogleScholarGoogle Scholar | 20421942PubMed |
[8] Fraser, J.A. et al. (2005) Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437, 1360–1364.
| Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFCrur3E&md5=a456eedea786bd45b45415aacdd02210CAS | 16222245PubMed |
[9] Meyer, W. et al. (2009) Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47, 561–570.
| Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht1agu7bF&md5=a2726b60fc37f15cc3c6d2652fe8a686CAS | 19462334PubMed |
[10] Hagen, F. et al. (2013) Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS ONE 8, e71148.
| Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtlajs7bK&md5=78c8c5a8e8f87069b6a8d74899d46c0cCAS | 23940707PubMed |
[11] Kaocharoen, S. et al. (2013) Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Negl. Trop. Dis. 7, e2297.
| Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand.Crossref | GoogleScholarGoogle Scholar | 23861989PubMed |
[12] Engelthaler, D.M. et al. (2014) Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio 5, e01464-14.
| Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal.Crossref | GoogleScholarGoogle Scholar | 25028429PubMed |
[13] Billmyre, R.B. et al. (2014) Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio 5, e01494-14.
| Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution.Crossref | GoogleScholarGoogle Scholar | 25073643PubMed |
[14] Lizarazo, J. et al. (2014) Retrospective study of the epidemiology and clinical manifestations of Cryptococcus gattii infections in Colombia from 1997-2011. PLoS Negl. Trop. Dis. 8, e3272.
| Retrospective study of the epidemiology and clinical manifestations of Cryptococcus gattii infections in Colombia from 1997-2011.Crossref | GoogleScholarGoogle Scholar | 25411779PubMed |
[15] Trilles, L. et al. (2008) Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Mem. Inst. Oswaldo Cruz 103, 455–462.
| Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsVKjtLrI&md5=ce81570037cd514f7b3e3a936b42162eCAS | 18797758PubMed |
Biographies
Carolina Firacative was recently awarded a PhD from The University of Sydney and is now a postdoctoral fellow at the Molecular Mycology Research Laboratory, Westmead Millennium Institute. Her research focuses on the phenotypic and genotypic characterisation of clinically important fungal pathogens.
Dr Kennio Ferreira-Paim is a Post-doctoral fellow in the Molecular Mycology Research Laboratory at the Center for Infectious Diseases and Microbiology, Westmead Millennium Institute and a Biomedical Scientist at the Clinical Hospital of the Triangulo Mineiro Federal University, Uberaba, Brazil where he recently concluded his PhD in Tropical Medicine and Infectious Diseases. His research focuses on the molecular epidemiology of Cryptococcus spp. and studying the molecular basis of fungal virulence using gene knockout and reconstitution and animal virulence models. He is CAPES Science without borders visiting fellow (#9313133) from Brazil.
Dr Luciana Trilles is a Medical Mycologist working as researcher and curator of the Culture Collection of Pathogenic Fungi and professor in the Infectious Diseases Post-Graduation Course at the Infectious Diseases Institute, Fundação Oswaldo Cruz (FIOCRUZ), Rio de Janeiro, Brazil. Her research focuses on ecology, molecular epidemiology, molecular diagnosis and identification of systemic mycosis’ agents.
Dr David M Engelthaler is the Director of TGen North, part of the non-profit Translational Genomics Research Institute, in Flagstaff, AZ, USA. David has over 20 years of public health research and practice history and was previously the Arizona State Epidemiologist and a biologist for the U.S. Centers for Disease Control and Prevention. He has published numerous papers on epidemiology, disease ecology, genetics, and microbiology.
Professor Wieland Meyer is a Molecular Medical Mycologist and academic at the Sydney Medical School, The University of Sydney and the Fundação Oswaldo Cruz (FIOCRUZ) in Rio de Janeiro, Brazil, heading the Molecular Mycology Research Laboratory within the Centre for Infectious Diseases and Microbiology, Westmead Millennium Institute. His research focuses on phylogeny, molecular identification, population genetics, molecular epidemiology and virulence mechanisms of human and animal pathogenic fungi. He is the Convener of the Mycology Interest Group of ASM, the Vice-President of the International Society of Human and Animal Mycology (ISHAM) and a member of the Executive Committee of the International Mycological Association (IMA).