The Westmead Medical Mycology Collection: basis for research and diagnosis of fungal diseases
Wieland Meyer A C , Krystyna Maszewska A , Aziza Khan A and Kennio Ferreira-Paim A BA Molecular Mycology Research Laboratory, Centre for Infectious Diseases and Microbiology, Sydney Medical School – Westmead Hospital, Marie Bashir Institute for Infectious Diseases and Biosecurity, The University of Sydney, Westmead Millennium Institute for Medical Research, Sydney, Australia
B Infectious Disease Department, Triangulo Mineiro Federal University, Uberaba, Minas Gerais, Brazil
C Corresponding author. Tel: +61 2 8627 3430, Fax: +61 2 9891 5317, Email: wieland.meyer@sydney.edu.au
Microbiology Australia 36(2) 60-63 https://doi.org/10.1071/MA15021
Published: 17 March 2015
The Westmead Medical Mycology Collection is completing 20 years of existence. During this time there have been 10,073 strains deposited representing 437 species, which are currently maintained in the collection. Established originally under the curation of Professor Wieland Meyer at the Molecular Mycology Research Laboratory, in the Centre for Infectious Diseases and Microbiology at the Sydney Medical School – Westmead Hospital, The University of Sydney, it recently moved to the new Westmead Millennium Institute for Medical Research in Westmead, Australia. Its primary aim is to preserve Australian human and animal pathogenic fungal biodiversity while providing reference and clinical strains for the mycology community. The stored strains are identified phenotypically, biochemically and molecularly. They are stored either lyophilised, in glycerol at −80°C or as living culture at 14°C. The majority of the stored strains are the result of specific clinical, molecular epidemiological and basic science projects. As such, the pathogenic yeasts Cryptococcus neoformans and C. gattii account for 54% of the specimens deposited. To further characterise the maintained strains specific MultiLocus Sequence Typing schemes have been developed for C. neoformans, C. gattii, Scedosporium apiospermum, S. aurantiacum, S. boydii and Pneumocystis jirovecii, which are publically accessible at http://mlst.mycologylab.org. The collection also formed the basis for the development of the quality controlled ISHAM-ITS sequence database for human and animal pathogenic fungi accessible at http://its.mycologylab.org.
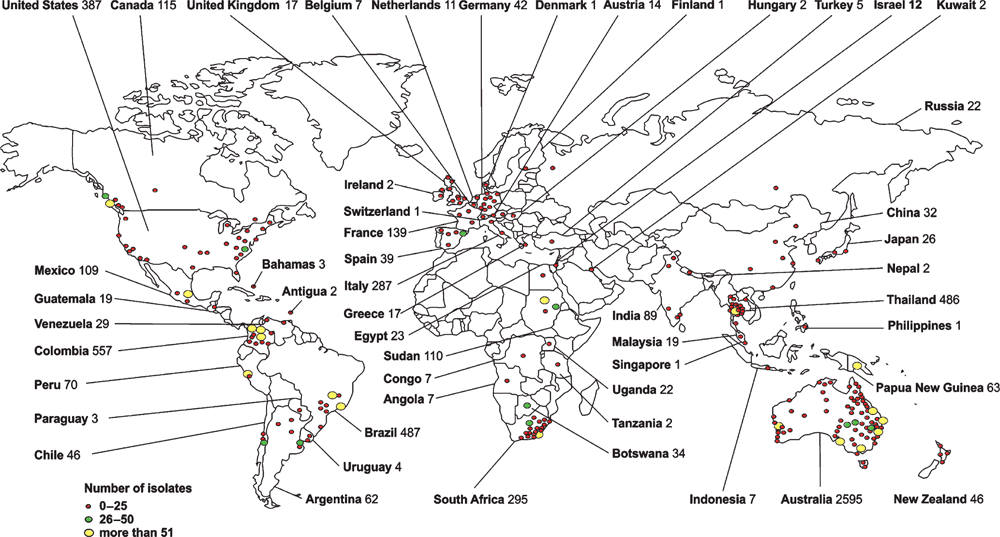
The Westmead Medical Mycology Collection (WM culture collection)
The storage of biological specimens is crucial for the preservation of microbial biodiversity and offering to the scientific community a wide range of data that can be used for diagnostic comparisons, as well as clinical and basic research. During the past 20 years, the WM Culture Collection has been focused on the culturing and storage of fungal strains, collecting 10,073 strains, representing 425 human and animal pathogenic fungal species, isolated from clinical, veterinary and environmental sources, from 52 countries (Figure 1). The collection maintains 134 type cultures and all reference strains for the major molecular types of the C. neoformans/C. gattii species complex1. The strains are characterised by applying traditional phenotypic, biochemical and advanced molecular techniques. The collection was originally established under the curation of Professor Wieland Meyer at the Molecular Mycology Research Laboratory (MMRL) in 1995 based on an existing strain collection started by Professor Tania Sorrell at the Centre for Infectious Diseases and Microbiology (CIDM) at Sydney Medical School–Westmead Hospital, The University of Sydney. With the move of CIDM and MMRL to the new Westmead Millennium Institute for Medical Research in June 2014 the collection found also a new permanent home. The collection maintains strains from a number of national and international studies, including the Australian Cryptococcus studies, the Australian candidemia study, the Australian Scedosporium study and the Latin American and Brazilian cryptococcosis studies. The collection has close collaborations with other national culture collections at the SA Pathology, Adelaide, the Royal North Shore Hospital, Sydney, St Vincent Hospital, Sydney, Veterinary Pathology at the Faculty of Veterinary Sciences at Sydney University, Sydney and the PathWest – QEII Medical Centre, Perth and international collections in Austria, Argentina, Brazil, Chile, Colombia, France, Germany, Greece, Italy, Japan, Malaysia, Mexico, New Zealand, Peru, Portugal, Spain, South Africa, Thailand, Taiwan, The Netherlands and the USA. The collection is part of the Australian Microbial Resources Research Network (AMRRN), the Australian Microbial Resources Information Network (AMRiN) (http://amrin.ala.org.au/), the Council of Heads of Australian Collections of Microorganisms (CHACM), and of the Atlas of Living Australia (http://www.ala.org.au).
Strain storage
After samples are received, they are identified using phenotypic or biochemical methods and then a single yeast colony is selected to be subcultured on Sabouraud dextrose agar plates for 48 hours at 30°C and/or 37°C for DNA extraction and preparation for long-term storage. A loop of the strain is mixed with skimmed milk, inoculated in sterilised glass vials and then processed in the Alpha 1–4 LSC Freeze Dryer®. All samples are stored either freeze dried or at –80°C. Filamentous fungi are morphologically identified and then subcultured on Sabouraud dextrose agar for 48 hours at 20°C for DNA extraction and preparation for long-term storage. A 4 cm2 section of the media is removed and inoculated in sterile glass vials containing 1 mL of sterile water, labelled and stored at 20°C. Metadata of the strains are stored electronically using the software package BioloMICS (www.bio-aware.com, Hannut, Belgium), that keeps track of the number of stock in the collection and manages new strains using the stock management system.
The collection and molecular epidemiological studies
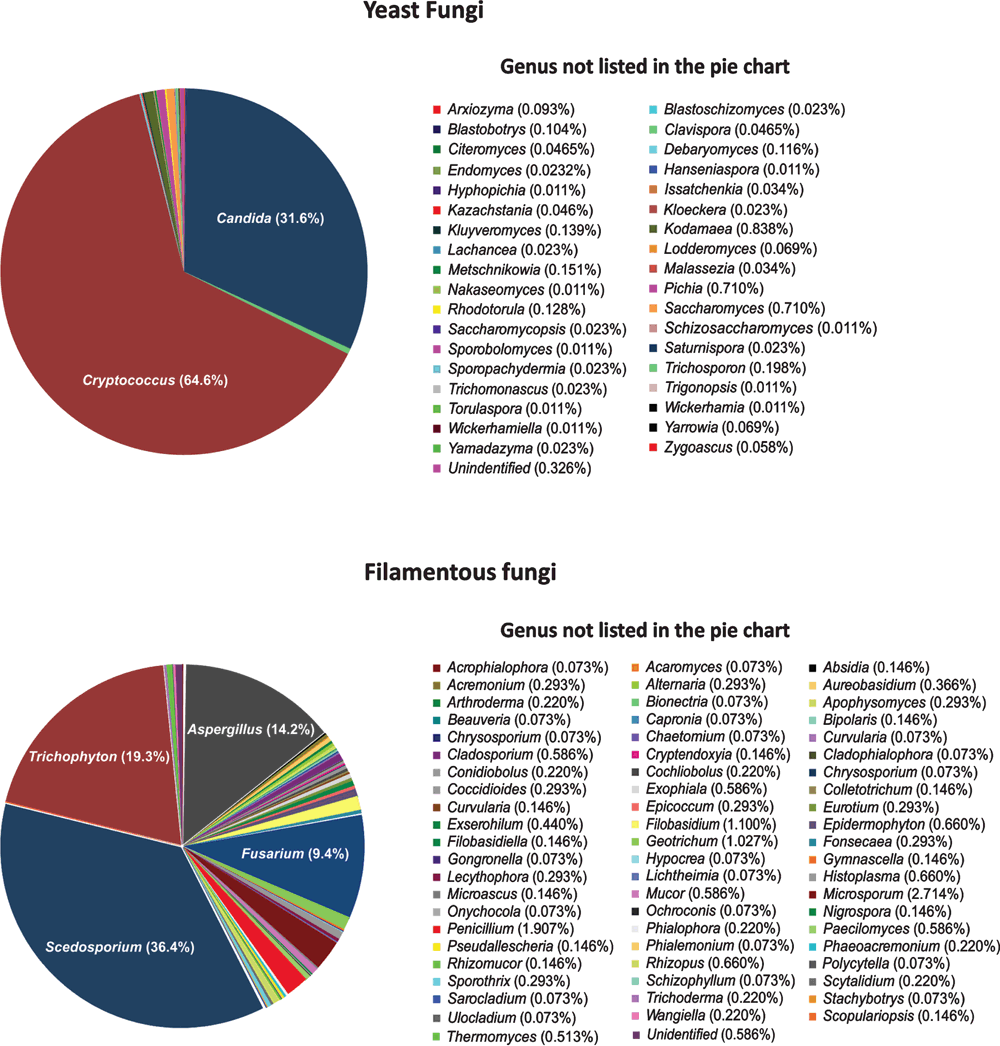
The genus Cryptococcus accounts for the highest number of samples (54%), with C. neoformans and C. gattii representing 80.5% of those species (Figure 2). This reflects one of the major research themes of the MMRL to understand the molecular epidemiology and virulence profiles of the etiological agents of cryptococcosis, considered one of the most common invasive fungal diseases in humans and responsible for more than 1 million cases per year and around 650,000 deaths in sub-Saharan Africa2. Within the 5,465 cryptococcal isolates available in the WM culture collection, 618 strains are typed using the International Society for Human and Animal Mycology (ISHAM) MultiLocus Sequencing Typing (MLST) consensus scheme1 (http://mlst.mycologylab.org) and whole genome sequencing was performed for 119 strains3 (Meyer and Firacative, unpublished data). All C. neoformans major molecular types (VNI, VNII, VNB, and VNIV) and 56 out of 324 sequence types (ST) currently described are present in the WM culture collection. All C. gattii major molecular types (VGI, VGII, VGIII, and VGIV) and 118 from the 336 STs described for C. gattii are available in the WM culture collection. Candida isolates represent the second most representative genus within the WM culture collection (122 species and 2,719 isolates), several of them were typed by molecular techniques. Filamentous and dimorphic fungi, especially those described as human pathogens including Aspergillus, Fusarium, Penicilium, Pseudallescheria, Fonsecaea and Histoplasma, are deposited as well. Among the filamentous fungi maintained in the collection the major pathogenic Scedosporium spp., opportunistic agents involved in pulmonary infections accounting for 497 strains (Figure 2), for which also specific MLST schemes, containing five genes4 have been developed, and are available at http://mlst.mycologylab.org, to enable a global molecular epidemiology survey of S. apiospermum, S. aurantiacum, and S. boydii.
|
The collection and molecular identification of fungal species
Since fungal identification and taxonomy has remarkably improved during the last decade several recognised species such as Fusarium solani, Paracoccidioides brasiliensis, and Sporothrix schenckii have been distinguished as complexes of cryptic species. In this context, the sequencing of the ribosomal regions, such as the Internal Transcribed Spacer (ITS), has been used for fungi identification for more than 10 years5. The ITS region was shown to be the most variable region within the ribosomal locus, being able to distinguish most closely related species, and as such has been selected as the universal fungal DNA barcode in 20126. It has been used frequently for phylogenetic studies5–7 and in the Assembling the Fungal Tree of Life (AFTOL) projects (http://tolweb.org). The WM culture collection formed the core unit of a global mycology research network combining 14 leading medical mycology laboratories, to establish the first quality controlled ITS database, the ISHAM-ITS reference database, which is available either via the ISHAM website at http://www.isham.org/ or directly at http://its.mycologylab.org. The database is constantly extended and accounts now for more than 3,000 sequences. More than 900 isolates from the WM culture collection are reference strains for the ISHAM-ITS database, 28.8% and 20.4% being Cryptococcus spp. and Candida spp., respectively. The remaining isolates represent the diversity of the fungal kingdom and etiological agents of mucormycosis, such as Lichtheimia corymbifera and invasive fungal infections, such as the rare Blastobotrys proliferans. The WM culture collection formed the basis for the development of a large number of molecular and MALDI-TOF based identification methods for human and animal pathogenic fungi, including: pan-fungal PCR8, genus/species-specific PCR9, real-time PCR10, reverse line blots11, rolling circle amplification12 and MALDI-TOF13.
The general mission of the WM culture collection is to continue to preserve and provide the mycology community with the Australian and global biodiversity of human and animal pathogenic fungi and associated metadata for clinical and basic research at a national and international level.
Acknowledgements
We thank all the members of the Australian and global mycology community that placed their trust in the WM culture collection and contributed to the construction of this important resource of biological diversity of human and animal pathogenic fungi.
References
[1] Meyer, W. et al. (2009) Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47, 561–570.| Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht1agu7bF&md5=a2726b60fc37f15cc3c6d2652fe8a686CAS | 19462334PubMed |
[2] Perfect, J.R. (2013) Efficiently killing a sugar-coated yeast. N. Engl. J. Med. 368, 1354–1356.
| Efficiently killing a sugar-coated yeast.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXls1aktLY%3D&md5=031f89c3da4921434143075e12c69249CAS | 23550675PubMed |
[3] Engelthaler, D.M. et al. (2014) Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio 5, e01464-14.
| Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal.Crossref | GoogleScholarGoogle Scholar | 25028429PubMed |
[4] Bernhardt, A. et al. (2013) Multilocus sequence typing of Scedosporium apiospermum and Pseudallescheria boydii isolates from cystic fibrosis patients. J. Cyst. Fibros. 12, 592–598.
| Multilocus sequence typing of Scedosporium apiospermum and Pseudallescheria boydii isolates from cystic fibrosis patients.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXpt1Cmtr4%3D&md5=57ee68d4fa6e57a93f56343f12579b68CAS | 23764085PubMed |
[5] Fell, J.W. et al. (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50, 1351–1371.
| Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXktF2gt7Y%3D&md5=e41d3e9da1e4cacc8ed56d824c51fc85CAS | 10843082PubMed |
[6] Schoch, C.L. et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246.
| Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xmt12mu7o%3D&md5=9f59bbfaa2c143e572b541e36ee6653eCAS | 22454494PubMed |
[7] Ferreira-Paim, K. et al. (2014) Phylogenetic analysis of phenotypically characterized Cryptococcus laurentii isolates reveals high frequency of cryptic species. PLoS ONE 9, e108633.
| Phylogenetic analysis of phenotypically characterized Cryptococcus laurentii isolates reveals high frequency of cryptic species.Crossref | GoogleScholarGoogle Scholar | 25251413PubMed |
[8] Lau, A. et al. (2007) Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J. Clin. Microbiol. 45, 380–385.
| Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXjtFGnsL4%3D&md5=cda2d024aa01389cd360fe53e9793457CAS | 17122000PubMed |
[9] Harun, A. et al. (2011) Development and validation of a multiplex PCR for detection of Scedosporium spp. in respiratory tract specimens from patients with cystic fibrosis. J. Clin. Microbiol. 49, 1508–1512.
| Development and validation of a multiplex PCR for detection of Scedosporium spp. in respiratory tract specimens from patients with cystic fibrosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFKrtL7M&md5=7de1d3a031ed8a068ca3a265fabd74e3CAS | 21325557PubMed |
[10] Lau, A. et al. (2008) Multiplex tandem PCR: a novel platform for rapid detection and identification of fungal pathogens from blood culture specimens. J. Clin. Microbiol. 46, 3021–3027.
| Multiplex tandem PCR: a novel platform for rapid detection and identification of fungal pathogens from blood culture specimens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFOku7vN&md5=db6371fdc74b7594dd5656addb41534fCAS | 18632914PubMed |
[11] Zeng, X. et al. (2007) Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens. J. Clin. Microbiol. 45, 2872–2880.
| Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtFemtLzP&md5=a046598c6d33e86a2bb0282f350357fdCAS | 17634313PubMed |
[12] Trilles, L. et al. (2014) Identification of the major molecular types of Cryptococcus neoformans and C. gattii by Hyperbranched rolling circle amplification. PLoS ONE 9, e94648.
| Identification of the major molecular types of Cryptococcus neoformans and C. gattii by Hyperbranched rolling circle amplification.Crossref | GoogleScholarGoogle Scholar | 24736745PubMed |
[13] Firacative, C. et al. (2012) MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS ONE 7, e37566.
| MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xot12rsb0%3D&md5=e5322f8ac1d839376fb3ab803f9bbbc4CAS | 22666368PubMed |
Biographies
The biography for Professor Wieland Meyer is on page 48.
Krystyna Maszewska is a research assistant at the MMRL who has graduated in Poland. She is managing the WM culture collection and carries out molecular identification using ITS1/2 and D1/D2 sequencing and genotyping of pathogenic fungi using PCR-fingerprinting, URA5-RFLP and MLST.
Aziza Khan is a research assistant at the MMRL who has completed her MSc in Medicine, with a focus on Infectious Diseases and Immunology at the University of Sydney. She performs ITS1/2 sequencing for the identification of pathogenic fungi for the development of the ISHAM-ITS database and conducts sequences for the selection of potential alternative DNA barcodes. She is working on the stability of Cryptococcus hybrid strains and conducts virulence studies for of various human pathogenic fungi using mice and Galleria mellonella larvae.
Kennio Ferreira-Paim is a Post-doctoral fellow in the MMRL at the CIDM, Westmead Millennium Institute and a Biomedical Scientist at the Clinical Hospital of the Triangulo Mineiro Federal University, in Uberaba, Brazil where he recently concluded his PhD in Tropical Medicine and Infectious Disease. His research focuses on the molecular epidemiology of Cryptococcus spp. and studying the molecular basis of fungal virulence using gene knockout and reconstitution and animal virulence models. He is a CAPES Science without borders visiting fellow (#9313133) from Brazil.