DNA barcoding of human and animal pathogenic fungi: the ISHAM-ITS database
Laszlo Irinyi A and Wieland Meyer A BA Molecular Mycology Research Laboratory, Centre for Infectious Diseases and Microbiology, Sydney Medical School – Westmead Hospital, Marie Bashir Institute for Infectious Diseases and Biosecurity, The University of Sydney, Westmead Millennium Institute, 176 Hawkesbury Road, Westmead, Sydney, NSW 2145, Australia
B Tel: +61 2 8627 3430, Fax: +61 2 9891 5317, Email: wieland.meyer@sydney.edu.au
Microbiology Australia 36(2) 44-48 https://doi.org/10.1071/MA15017
Published: 16 March 2015
Human and animal fungal pathogens are a growing threat worldwide. They lead to emerging infections and create new risks for established ones. As such, there is a growing need for the rapid and accurate identification of mycoses agents to enable early diagnosis and targeted antifungal therapy. An international consortium of medical mycology laboratories was formed in order to establish a quality controlled ITS database under the umbrella of the ISHAM (International Society for Human and Animal Mycology) working group on ‘DNA barcoding of human and animal pathogenic fungi’. The new database provides the medical community with a freely accessible tool via http://www.isham.org/ or directly at http://its.mycologylab.org/ to rapidly and reliably identify most mycoses agents. The average intra-species variation of the ITS sequences currently included in the database ranges from 0 to 2.25%, highlighting the fact, that the ITS region on its own is insufficient for the reliable identification of certain pathogenic fungal species.
The number of human and animal mycoses, ranging from superficial to invasive fungal infections, has significantly increased over the past three decades, causing serious public health burdens and increased risk of biodiversity loss among animal species1,2. To better understand, control and treat fungal infections, more rapid and accurate identification of the causal agents is essential. Traditional identification based on morphology and biochemistry is time-consuming and requires morphological and taxonomical expertise. To overcome these limitations, DNA barcoding offers a practical approach for species identification, which is less demanding in terms of taxonomical expertise. DNA barcoding consists of using short sequences (500–800 bp) for the identification of organisms at species level by comparison to a reference collection of well-identified species. The concept of barcoding is that species identification must be accurate, fast, cost-effective, culture independent, universally accessible and feasible for non-experts. The principles of barcoding are that: (i) interspecies variation should exceed intraspecies variation, to create a barcode gap3, and (ii) identification is straightforward, when a sequence is unique to a single species and constant within each species4.
ITS as the current official DNA barcode for fungi
The current official fungal DNA barcode5, the internal transcribed spacer (ITS) region has long been used in molecular identification and phylogenetic studies of human and animal pathogenic fungi. The ITS region is easy to amplify with universal primers suitable for most fungal species and show sufficient genetic variability at interspecies level. The ITS sequences are used routinely by the medical community for fungal identification at the species level on the basis of matching sequences in publicly accessible databases, such as GenBank. However, its wide-spread applicability is still limited by the absence of quality-controlled reference databases. According to a recent study, 10% of the publicly available fungal ITS sequences were identified incorrectly at species level. Many of the ITS sequences deposited in public databases are incomplete or wrongly annotated6. Moreover, no studies have been done to evaluate the ITS region as an official standard barcode in clinically relevant fungal species.
Establishment of the ISHAM-ITS reference database
To address these issues, a working group of the International Society for Human and Animal Mycology (ISHAM) on ‘Barcoding of Medical Fungi’ was established in 2011. The working group identified the necessity to: (i) generate a medical barcode database by incorporating existing fungal group-specific databases; (ii) extend the number of quality-controlled ITS sequences to cover all medically important fungal species; (iii) evaluate the value of ITS as a barcode at intra- and interspecies level, and (iv) eventually incorporate these sequences into the GenBank and other reference databases.
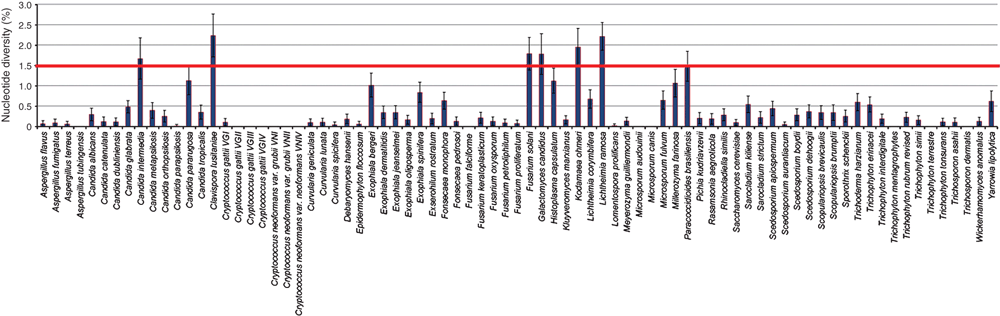
Fourteen mycology laboratories from three continents initially generated 3200 complete ITS sequences representing 524 clinically relevant species. The ISHAM-ITS reference database is freely accessible at http://its.mycologylab.org/ and http://www.isham.org/. It contains 226 species represented by one strain, 116 species by two strains, and 182 species by a minimum of three to a maximum of 115 sequences. The medically most relevant species are represented in the database by 20–115 strains. The lengths of complete ITS sequences in the ISHAM-ITS reference database range from 285 to 791 bp. The shortest complete ITS sequences are assigned to Candida haemulonis (285 bp), Clavispora lusitaniae (293 bp), and the longest ones to Candida glabrata (791 bp) and Lichtheimia ramosa (770 bp). The mean nucleotide length of ITS sequences in the database is 503 bp. The length, continuity and annotation of the ITS sequences have been checked using the software ITSx 1.0.77.
ITS intraspecies variation
The average intraspecies genetic diversity of the ITS region in medically relevant fungal species contained in the ISHAM-ITS database ranges between 0 and 2.25%, but in 170 species it is less than 1.5%. In 138 species it is less than 0.5%, in 27 species it ranges, between 0.5–1.0%, in five species (Exophiala bergeri, Millerozyma farinosa, Histoplasma capsulatum, Candida pararugosa and Paracoccidioides brasiliensis) between 1.01–1.5%, in four species (Candida intermedia, Galactomyces candidus, Fusarium solani and Kodamaea ohmeri) 1.5–2.0%, and in two species (Lichtheimia ramosa and Clavispora lusitaniae) it is more than 2% (Figure 1). The distribution of polymorphic sites revealed similar results. In 117 species, the number of polymorphic sites is less than five, in 35 species it is between five and ten, in 11 species between 11 and 15, in six species between 16 and 20 and finally more than 20 in seven species. The species with the highest number of segregating sites are Cryptococcus albidus (21 sites), the complex of F. solani (21 sites), C. lusitaniae (22 sites), Candida glabrata (22 sites), K. ohmeri (23 sites), H. capsulatum (38 sites) and L. ramosa (55 sites). Clinically important species have a low intraspecies variability in ITS regions making the ITS sequencing a useful genetic marker to be used for their identification. For the species with higher than 1.5% intraspecies diversity, additional molecular methods may be required for their reliable identification. Previous studies have shown that the genetic diversity of the ITS regions in fungi varies between taxa and that a universal cut off value to delineate species cannot be established8. Intraspecies diversity in medical fungi may be due to intra-genomic polymorphisms.
ITS interspecies variation
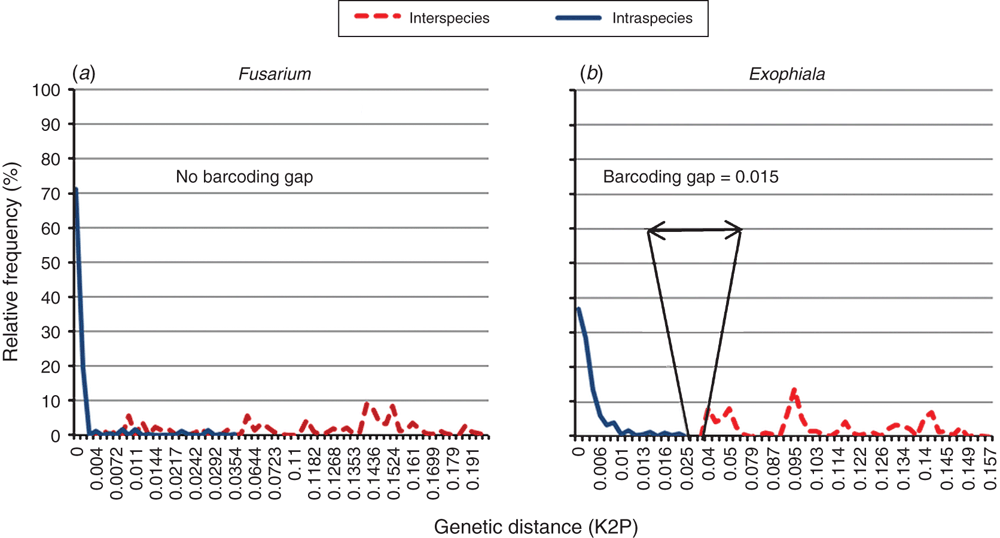
In 13 taxa, sharing the same phylogenetic clades, a clear barcoding gap (K2P9 distance) was detected. This means that the highest intraspecies distances were smaller than the lowest genetic distances between species, generating a ‘barcoding gap’. An example of taxa with and without barcoding gap is shown on Figure 2. The smallest barcoding gap (0.0002) exists in the Microsporum spp., while the largest one is present in the Cladophialophora spp. (0.09). However, four taxa have no clear barcoding gap: Cryptococcus, Fusarium, Scedosporium and Trichophyton. In these taxa, the correct identification to the species level may be problematic when only using the ITS region as a genetic marker. As such, additional genetic markers and/or molecular methods are required.
Linking the ISHAM-ITS database to GenBank and UNITE
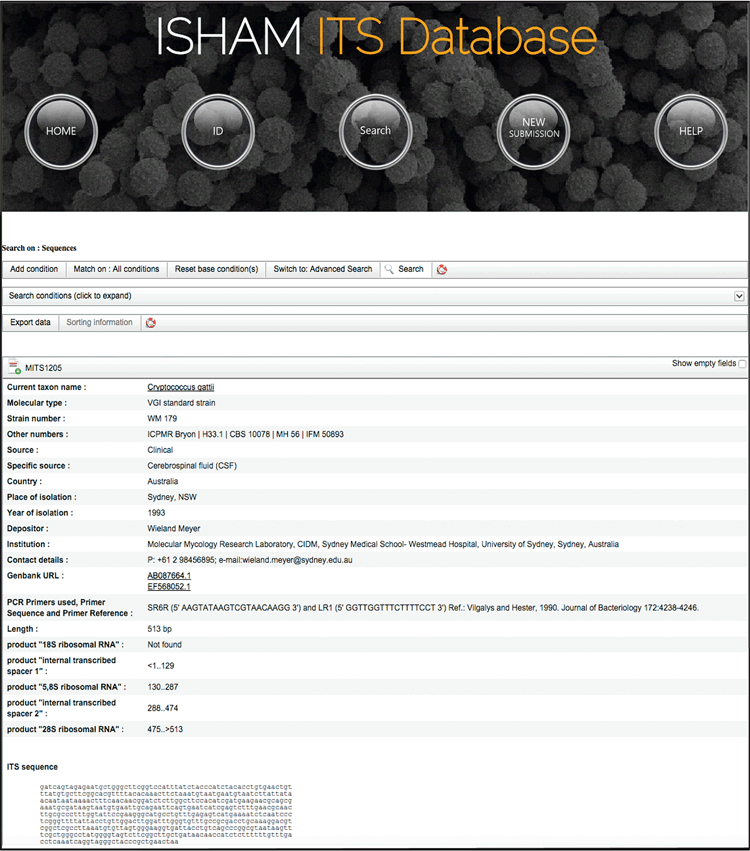
As a result of the collaboration with NCBI, all sequences are submitted to GenBank where they are labelled specifically, indicating that they are part of the ISHAM-ITS database and that they are quality controlled sequences. The definition line of each ITS sequence submission in GenBank covers the current taxon name of the species, the original strain number and a unique ‘ISHAM-ITS ID’ identifier (e.g. MITS1; MITS2….) as follows: ‘Acremonium acutatum strain FMR 10368 isolate ISHAM-ITS_ID MITS1 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence’. Following a BLAST search in GenBank, the user can clearly identify the query sequence selecting the ISHAM-ITS record from the Blast hits list. In GenBank, each ISHAM-ITS record is directly linked to the ISHAM-ITS database where more metadata are available for the associated strain (Figure 3). Moreover, sequences selected from the ISHAM-ITS database expand the number of medically relevant species represented in the RefSeq Targeted Loci (RTL) ITS reference database at NCBI10. Of the 421 fungal species contained in the ISHAM-ITS database, 71 are represented by Type cultures and have been submitted to RTL at NCBI. Conversely, 281 RefSeq sequences representing Type and verified material have been added to the ISHAM-ITS database. The NCBI and ISHAM curators are working together to update the species names in response to ongoing taxonomy and nomenclatural changes. In addition to GenBank, the sequences are also submitted to the UNITE database11, where they are specifically labelled and directly linked to the ISHAM-ITS reference database.
|
Value of the ITS as a fungal DNA barcode
Taking the current data into account, most of the medically relevant fungal species can be identified based on their ITS region, verifying its status as a primary standard DNA barcode for fungi. However, in some cases the ITS has limitations in differentiating species. There are two possible reasons for this: either the taxa are insufficiently studied or the ITS region is simply an inappropriate marker for discrimination between closely related species. To overcome these limitations alternative loci and/or molecular methods are required. The occurrence of taxa without a barcoding gap may also be explained by the fact that the algorithms used by the barcoding community to calculate the genetic distances (K2P) or the algorithm used in BLAST12 for pairwise sequence matching between the query sequence and reference sequences represent different approaches from those commonly used for phylogenetic analyses.
The ISHAM-ITS database is intended to cover all clinically relevant fungal species. It is open for further sequence submission to expand coverage of medically relevant species with a sufficient number of strains, either via direct submission through the database or by contacting the curators of the database at: laszlo.irinyi@sydney.edu.au or wieland.meyer@sydney.edu.au.
Acknowledgements
The authors thank all contributors to the ISHAM-ITS database (http://its.mycologylab.org). This work was supported by a NHMRC grant #APP1031952 to WM.
References
[1] Brown, G.D. et al. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv13.| Hidden killers: human fungal infections.Crossref | GoogleScholarGoogle Scholar | 23253612PubMed |
[2] Fisher, M.C. et al. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194.
| Emerging fungal threats to animal, plant and ecosystem health.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmtVeqtb4%3D&md5=a8e99bd4c6ab5772af7faf09cb455b9cCAS | 22498624PubMed |
[3] Meyer, C.P. and Paulay, G. (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 3, e422.
| DNA barcoding: error rates based on comprehensive sampling.Crossref | GoogleScholarGoogle Scholar | 16336051PubMed |
[4] Hebert, P.D. et al. (2003) Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321.
| Biological identifications through DNA barcodes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXktVWiu7g%3D&md5=2e5a20ae9085df0d761a645324195c6eCAS | 12614582PubMed |
[5] Schoch, C.L. et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246.
| Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xmt12mu7o%3D&md5=9f59bbfaa2c143e572b541e36ee6653eCAS | 22454494PubMed |
[6] Nilsson, R.H. et al. (2006) Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1, e59.
| Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective.Crossref | GoogleScholarGoogle Scholar | 17183689PubMed |
[7] Bengtsson-Palme, J. et al. (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919.
| Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data.Crossref | GoogleScholarGoogle Scholar |
[8] Nilsson, R.H. et al. (2008) Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. Online 4, 193–201.
| 19204817PubMed |
[9] Kimura, M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
| A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3MXmtFSktg%3D%3D&md5=ec9402a8d1a23908437333c41961a2b6CAS | 7463489PubMed |
[10] Schoch, C.L. et al. (2014) Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database (Oxford) 2014, bau061.
| Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi.Crossref | GoogleScholarGoogle Scholar | 24980130PubMed |
[11] Kõljalg, U. et al. (2013) Towards a unified paradigm for sequence-based identification of Fungi. Mol. Ecol. 22, 5271–5277.
| Towards a unified paradigm for sequence-based identification of Fungi.Crossref | GoogleScholarGoogle Scholar | 24112409PubMed |
[12] Altschul, S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
| Basic local alignment search tool.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXitVGmsA%3D%3D&md5=d2bbcc65aaf1fb0004b58c7099f4eb5cCAS | 2231712PubMed |
Biographies
Laszlo Irinyi is a Post-doctoral fellow in the Molecular Mycology Research Laboratory at the Center for Infectious Diseases and Microbiology, Westmead Millennium Institute. He completed his PhD on the phylogeny of Didymellaceae at University of Debrecen, Hungary. His research focuses on barcoding and molecular identification of human and animal pathogenic fungi. He is the curator of the International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database.
Professor Wieland Meyer is a Molecular Medical Mycologist and academic at the Sydney Medical School, The University of Sydney and the Fundação Oswaldo Cruz (FIOCRUZ) in Rio de Janeiro, Brazil, heading the Molecular Mycology Research Laboratory within the Centre for Infectious Diseases and Microbiology, Westmead Millennium Institute. His research focuses on phylogeny, molecular identification, population genetics, molecular epidemiology and virulence mechanisms of human and animal pathogenic fungi. He is the Convener of the Mycology Interest Group of ASM, the Vice-President of the International Society of Human and Animal Mycology (ISHAM) and a member of the Executive Committee of the International Mycological Association (IMA).