Relative abundance of Mycobacterium in ovine Johne’s disease
Andy O Leu A , Paul Pavli B C , David M Gordon A , Jeff Cave D , Jacek M Gowzdz E , Nick Linden D , Grant Rawlin E , Gwen E Allison A C and Claire L O’Brien A B C FA Research School of Biology, Australian National University, Acton, ACT 0200, Australia
B Gastroenterology and Hepatology Unit, Canberra Hospital, Garran, ACT 2605, Australia
C Medical School, Australian National University, Acton, ACT 0200, Australia
D Department of Environment and Primary Industries, Wodonga, Vic. 3690, Australia
E AgriBio, Bundoora, Vic. 3083, Australia
F Corresponding author. Tel: +61 2 6244 4023, Email: claire.obrien@anu.edu.au
Microbiology Australia 36(1) 32-36 https://doi.org/10.1071/MA15010
Published: 6 March 2015
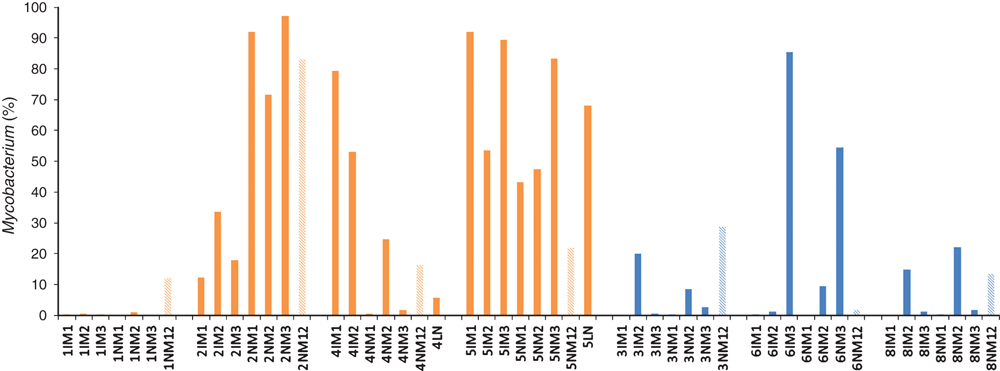
No study has determined what proportion of the total microbiota comprises the genus Mycobacterium in ovine Johne’s disease (OJD) tissues. We aimed to assess the relative abundance of Mycobacterium in the ileocaecal lymph node, involved and uninvolved ileal mucosa from sheep with and without OJD, using three extraction methods. Eight sheep, four with and four without OJD, were recruited. Pyrosequencing of the 16Sr RNA gene amplicons for all samples revealed that Mycobacterium represented between 0-92% (average 38%) of the total microbiota of samples from sheep with OJD, and 0-85% (average 13%) of sheep without OJD. Only sheep with OJD had samples that were positive for the IS900 (MAP) element. Mycobacterial strains other than MAP may provide competitive exclusion of MAP and should be further investigated.
Mycobacterium avium subspecies paratuberculosis (MAP) is the etiologic agent of ovine Johne’s disease (OJD), a chronic, contagious granulomatous enteritis of ruminants. The disease causes significant morbidity and mortality worldwide, resulting in significant economic losses1.
To our knowledge, no study has determined what fraction of the microbial community of non-OJD and OJD affected tissue comprises Mycobacterium. Cheung et al. (2013)2 found that Mycobacterium tuberculosis represented less than 1% of the total microbial community of sputum samples of infected patients. Does Mycobacterium comprise such a small fraction of the microbial community in all infectious states and specimens? MAP is a putative trigger of Crohn’s disease in humans, and is detected in some studies but not others, including a recent study of ours3. Is this because it is not being detected?
While other studies have quantified Mycobacterium or MAP in clinical and subclinical specimens4,5, no study has determined what fraction of the microbial community comprises Mycobacterium in mucosa from sheep with and without OJD. The primary aim is to assess the relative abundance of Mycobacterium in the microbial communities of macroscopically normal bowel mucosa obtained >50 cm downstream the ileocaecal valve (sheep without OJD) or involved mucosa (sheep with OJD), involved (or normal for sheep without OJD) ileal mucosa adjacent the ileocaecal lymph node, and the ileocaecal lymph node of sheep with and without OJD.
Samples of involved and uninvolved ileal mucosa, and ileocaecal lymph nodes were collected from eight sheep, four with and four without OJD, from farms around Rutherglen, Australia. Sterile gloves and implements were used to extract the ileocaecal node before the bowel was opened, so that it did not come into contact with the skin or bowel microbiota. Total DNA was extracted from each sample using three different extraction methods. The first protocol followed the Qiagen DNeasy kit protocol and included an enzymatic lysis step and bead-beating step. The second and third extractions were the same as the first, except the second protocol included a freeze-thaw step, and the third protocol a boiling step. Technical replicates were included for the first protocol. All samples were amplified using barcoded primers targeting the universal bacterial16S rRNA gene, and pyrosequenced using a 454 Genome Sequencer FLX-Titanium platform, as previously described3. Sequences were analysed using Mothur6. IS900 PCR was used to determine whether or not Mycobacterium detected in a given sample was MAP, or not.
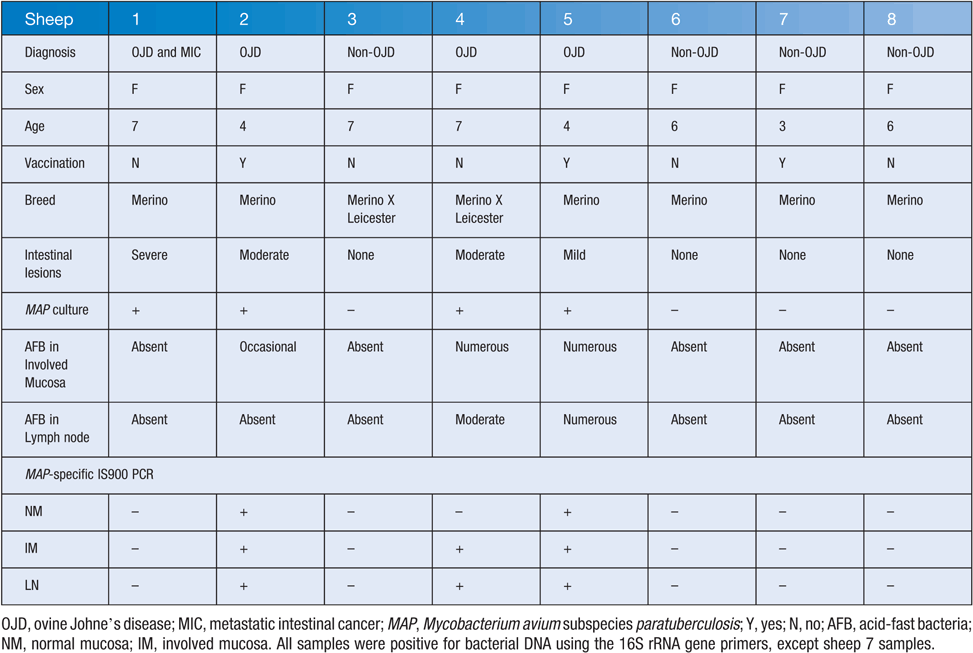
Four sheep (1, 2, 4 and 5; for example, see Figure 1) were diagnosed with OJD by qualified veterinarians and pathologists, using methods previously described7,8. Briefly, sheep were initially assessed as having OJD, or not, based on physical signs, including condition, scouring, ill-thrift, and lethargy during mobility. Diagnosis was confirmed by a NATA-accredited pathology laboratory (AgriBio, Victoria), and included MAP culture and DNA analysis, as well as microscopic visualization of acid-fast bacteria using ZN staining . Sheep 1 was initially suspected to have OJD, however negative culture and staining results excluded it from a diagnosis of OJD, and it was found to have metastatic cancer.

|
Pyrosequencing showed that Mycobacterium represented between 0–92% (average 38%) of the total microbiota of samples from sheep with OJD, and 0-85% (average 13%) of sheep without OJD (Figure 2). Mucosal samples from sheep 7 were excluded due to the inability to amplify products from them. Sheep 4 and 5 each had a positive lymph node sample for one extraction, comprising 6% and 68% Mycobacterium reads, respectively. These levels were similar to that observed in the mucosal samples from the same animal.
The lymph node and mucosa samples of sheep 1 were negative when tested with the MAP-specific primers. Sheep 1 may have had a paucibacillary form of OJD. Samples from all other sheep with OJD were positive, except the normal mucosa sample of sheep 4 (Table 1). All samples from sheep without OJD were negative for the IS900 element.
|
No significant effect of extraction method was detected for either Simpson or Shannon diversity estimates (Simpson Diversity, AOV: P > F = 0.384; Shannon Diversity, AOV: P > F = 0.443), nor technical replicates (Simpson Diversity, AOV: P > F = 0.398; Shannon Diversity, AOV: P > F = 0.566).
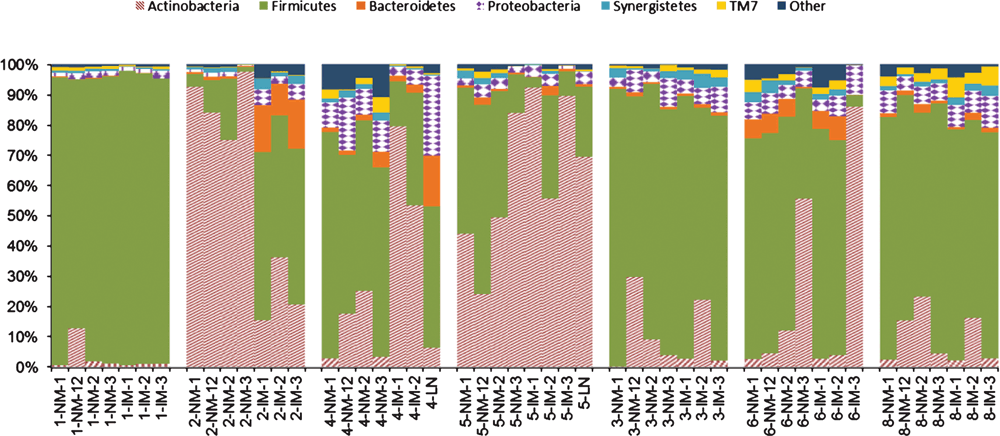
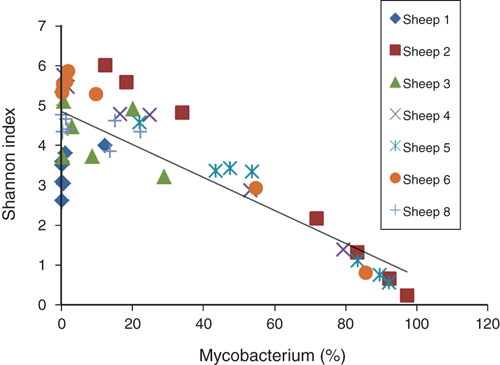
Each sample was covered by an average of 2, 644 quality sequences. Six phyla (Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Synergistetes, and TM7) represented ~98% of all sequences (Figure 3). Actinobacteria, of which Mycobacterium is a member, was overrepresented in sheep with OJD, except sheep 1. Microbial community diversity declined as Mycobacterium increased (R2=0.705, p=0.001) (Figure 4).
|
To our knowledge, this is the first study to describe the full microbial community of mucosa from sheep with and without OJD. Although studies have enumerated MAP in the feces of sheep using RT-PCR5,9, no study has determined what fraction of the microbial community Mycobacterium comprises, and whether or not these proportions differ in sheep with and without OJD, as well as various tissue types.
Bacterial translocation to the ileocaecal lymph node was observed in 50% (2/4) of sheep diagnosed with OJD. The microbial community of these nodes was similar to that of the involved and normal mucosa from the same sheep. Bacteria in the node are likely to be secondary invaders, present in the node due to a breakdown in the mucosal barrier. Using the same methods described here, we found that bacterial translocation in Crohn’s disease of humans is also non-specific. We also did not detect Mycobacterium in this study using 16S rRNA gene sequencing3. This suggests that Mycobacterium is unlikely to be a cause of Crohn’s disease.
Samples from sheep without OJD were not positive for MAP culture or the IS900 element, but other strains of Mycobacterium were present, representing on average 13% of the total microbiota. Strains of Mycobacterium other than MAP could occupy a niche in the sheep gastrointestinal tract that MAP would otherwise occupy, thereby protecting the animal from colonization of MAP. Future studies should assess the diversity of Mycobacterium strains associated with the mucosa of sheep with and without OJD.
We found the abundance of Mycobacterium, relative to other bacterial taxa in the mucosal samples, was significantly higher in sheep with OJD. Increases in Mycobacterium result in large decreases in Firmicutes. Members of the Firmicutes are important producers of short-chain fatty acids, which are shown to improve intestinal barrier function and repress inflammation10. Dysbiosis and reduced diversity would likely exacerbate symptoms of OJD, as beneficial bacteria are decreased and their roles in homeostasis unfulfilled.
Mycobacterium makes up a large fraction of the microbial communities of mucosa from sheep with OJD. Future studies should determine if MAP is the sole contributor to these levels of Mycobacterium in sheep with OJD, and assess the diversity of Mycobacterium in sheep with and without OJD. Other Mycobacterium strains may provide competitive exclusion of MAP.
Acknowledgements
We thank Greg Seymour for assisting in the collection of specimens.
References
[1] Salem, M. et al. (2013) Mycobacterium avium subspecies paratuberculosis: an insidious problem for the ruminant industry. Trop. Anim. Health Prod. 45, 351–366.| Mycobacterium avium subspecies paratuberculosis: an insidious problem for the ruminant industry.Crossref | GoogleScholarGoogle Scholar | 23054804PubMed |
[2] Cheung, M.K. et al. (2013) Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS ONE 8, e54574.
| Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXit1ChsLg%3D&md5=c24867699af12fecab1bae29f719b120CAS | 23365674PubMed |
[3] O’Brien, C.L. et al. (2014) Detection of bacterial DNA in lymph nodes of Crohn’s disease patients using high throughput sequencing. Gut 63, 1596–1606.
| Detection of bacterial DNA in lymph nodes of Crohn’s disease patients using high throughput sequencing.Crossref | GoogleScholarGoogle Scholar | 24429583PubMed |
[4] Park, K.T. et al. (2014) Development of a novel DNA extraction method for identification and quantification of Mycobacterium avium subsp. paratuberculosis from tissue samples by real-time PCR. J. Microbiol. Methods 99, 58–65.
| Development of a novel DNA extraction method for identification and quantification of Mycobacterium avium subsp. paratuberculosis from tissue samples by real-time PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXks1Wgtrs%3D&md5=7b1ece82390397de1004aa8abd2bafafCAS | 24534783PubMed |
[5] Logar, K. et al. (2012) Evaluation of combined high-efficiency DNA extraction and real-time PCR for detection of Mycobacterium avium subsp. paratuberculosis in subclinically infected dairy cattle: comparison with faecal culture, milk real-time PCR and milk ELISA. BMC Vet. Res. 8, 49.
| Evaluation of combined high-efficiency DNA extraction and real-time PCR for detection of Mycobacterium avium subsp. paratuberculosis in subclinically infected dairy cattle: comparison with faecal culture, milk real-time PCR and milk ELISA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVaktLnI&md5=746caea40c02dbbf19c742214ea008efCAS | 22551054PubMed |
[6] Schloss, P.D. et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541.
| Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXis1yltw%3D%3D&md5=2bd3a329be90d06651a79c07a2341d58CAS | 19801464PubMed |
[7] Gwozdz, J.M. et al. (2000) Vaccination against paratuberculosis of lambs already infected experimentally with Mycobacterium avium subspecies paratuberculosis. Aust. Vet. J. 78, 560–566.
| Vaccination against paratuberculosis of lambs already infected experimentally with Mycobacterium avium subspecies paratuberculosis.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3cvmt1Smtg%3D%3D&md5=c549ce8a8a7c6aaf7b3119603567f7e9CAS | 10979513PubMed |
[8] Whittington, R.J. et al. (2013) Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified Bactec 12B medium. J. Clin. Microbiol. 51, 3993–4000.
| Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified Bactec 12B medium.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXjtlWrsbs%3D&md5=1bf4eff20334a79f64159b42f83efbabCAS | 24048541PubMed |
[9] Sting, R. et al. (2014) Detection of Mycobacterium avium subsp. paratuberculosis in faeces using different procedures of pre-treatment for real-time PCR in comparison to culture. Vet. J. 199, 138–142.
| Detection of Mycobacterium avium subsp. paratuberculosis in faeces using different procedures of pre-treatment for real-time PCR in comparison to culture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvVKisbvP&md5=b6af46cbb4d2b2b4861831a39c009e7fCAS | 24280588PubMed |
[10] Pryde, S.E. et al. (2002) The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217, 133–139.
| The microbiology of butyrate formation in the human colon.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XptlCqs7g%3D&md5=22bb89ffcdf2b3809b3cb830f1d386eaCAS | 12480096PubMed |
Biographies
Andy Leu graduated from the Australian National University (ANU) with a Bachelor of Medical Science (Honours I) in 2013. His Honours project examined the relative abundance of Mycobacterium avium. paratuberculosis (MAP) in Johne’s disease tissues. Andy was recently awarded an Australian Postgraduate Award scholarship to pursue a PhD under the supervision of Dr Gene Tyson.
Paul Pavli is a gastroenterologist with clinical and basic scientific research interests in the inflammatory bowel diseases. He was on the steering committee of the multicentre Australian study using long-term anti-mycobacterial agents in Crohn’s disease (Selby et al., Gastroenterology, 2007). Current research interests are in the interaction between the innate inflammatory response and the intestinal microbiome.
David Gordon received his BSc from King’s College London and his PhD from McGill University, Montreal. His research concerns the ecology, population genetics and evolution of the Enterobacteriaceae, especially E. coli.
Jeff Cave graduated from Murdoch University in 1988 and spent two years in mixed private practice in South Australia before working in the Pacific for much of the 1990s. For the past 16 years he has worked as the District Veterinary Officer in Wodonga, Victoria.
Jacek Gwozdz is a Veterinary Pathologist at the State veterinary laboratory in Victoria. He received his PhD in Veterinary Science from the Massey University in New Zealand and worked over the past 16 years in research and diagnostic microbiology and pathology.
Nick Linden is a research scientist with the Victorian Department of Primary Industries and Environment, he has a particular interest in the lamb supply chain and is passionate about linking on farm production through to carcass appraisal and consumer acceptance. His recent projects have been focused on lamb feed conversion efficiency as well as on-line grading of lamb carcasses.
Grant Rawlin is Research Leader of Veterinary Pathobiology at the State veterinary laboratory in Victoria. His career has spanned veterinary diagnostics as a pathologist, research in veterinary and human microbiology and research into control of infectious diseases of animals at a population level.
Dr Gwen Allison received her PhD in Microbiology from North Carolina State University. Her research interests have included molecular biology of lactic acid bacteria and microbial ecology of the gastrointestinal tract and environment. The current research was conducted when Dr Allison was a Senior Lecturer at the ANU.
Dr Claire O’Brien received her PhD from the Australian National University Medical School (ANUMS) in 2012. Her main research interest is the gut microbiome, particularly in Crohn’s disease. Claire is an NHMRC Peter Doherty Fellow at the ANUMS and Canberra Hospital.