Use of Caenorhabditis elegans as a non-mammalian model system to study Candida virulence
Farkad Bantun A B , Sanjiveeni Dhamgaye A and Anton Y Peleg A C DA Department of Microbiology, Monash University, Clayton, Vic., Australia
B Department of Medical Laboratories Technology, Faculty of Applied Medical Sciences, Jazan University, Jazan, Saudi Arabia
C Department of Infectious Diseases, Central Clinical School, The Alfred Hospital and Monash University, Melbourne, Vic., Australia
D Corresponding author. Tel: +61 3 9076 8491, Fax: +61 3 9076 2431, Email: anton.peleg@monash.edu
Microbiology Australia 36(2) 98-100 https://doi.org/10.1071/MA15032
Published: 17 March 2015
Candida albicans forms part of the normal human commensal flora but has the ability to cause serious, invasive disease in those who are immunosuppressed. One of its key virulence determinants is its ability to transition from a yeast to a filamentous form. This article focuses on the utility of using the worm model, Caenorhabditis elegans, to study Candida pathogenesis. C. elegans provides an in vivo infection environment that is ideally suited to study the mechanisms of filamentation and its role in disease. Findings from the C. elegans-Candida model appear highly predictive of findings in a mammalian infection model.
C. albicans is one of the most common human fungal pathogens1. It is part of the human commensal flora, which colonizes gastrointestinal, mucocutanous, and genitourinary areas in nearly 80% of healthy individuals2. However, when host immune defenses are disrupted, severe invasive disease such as candidaemia can ensue3,4. One of the most important virulence factors that contribute to C. albicans pathogenesis is its ability to switch, reversibly, between yeast to filamentous (hyphal) forms3. The hyphal form of C. albicans is thought to be intricately related to its pathogenesis, with some studies showing that it is necessary for tissue destruction and invasion3,5–7.
C. elegans is a soil-dwelling nematode that has been used in biomedical science for over 30 years; however its use for studying microbial pathogenesis is more recent. C. elegans provides an excellent balance between complexity and logistic ease, as well as having significant ethical and financial advantages over mammalian infection models8–10. C. elegans has a fast generation time: it grows up to 1 mm in length and can produce genetically identical progeny in a 3-day life cycle11. The worm has been successfully used to uncover host immunity and virulence of pathogens similar to that implicated in human or other animal models during disease. C. elegans is a natural host to various pathogens including Microbacterium nematophilum, Drechmeria coniospora, Nematocida parisii, while it has also been used as model host for many pathogenic fungi including Cryptococcus neoformans12, Histoplasma capsulatum13, and most recently Penicillium marneffei14.
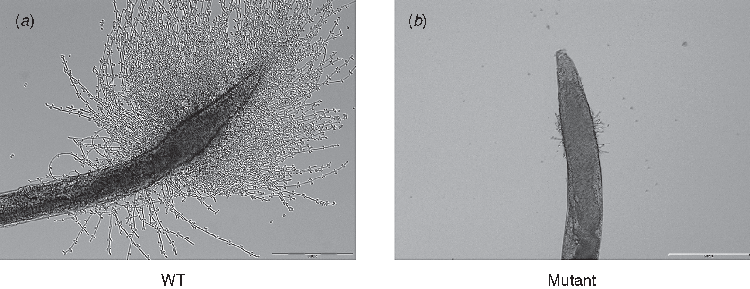
Although it has been challenging to find an appropriate non-mammalian model to study the role of filamentation in the pathogenesis of C. albicans, Breger and colleagues developed the C. elegans model to perform in vivo studies of C. albicans pathogenesis and antifungal compounds15. Using a standard feeding assay, C. albicans were ingested by adult C. elegans worms, and then worms were transferred into liquid media. C. albicans established a persistent intestinal tract infection and proliferated within the gut. The yeast cells would then undergo a morphological transition to form penetrative filaments, piercing the body of the worm and leading to its death. An example of this penetrative filamentation is shown in Figure 1A. In some ways, this has similarity to its proliferation in the human gastrointestinal tract and gut translocation to cause invasive disease15. Pukkila-Worley, Peleg and colleagues have also shown the successful use of C. elegans in elucidating C. albicans dimorphism7. By employing hyphal defective mutant strains (EFG1 and FLO8) we showed that hyphae formation is necessary for the pathogenesis of Candida as confirmed by attenuated virulence toward C. elegans. An example image of a worm infected with a hyphal-defective Candida mutant is shown in Figure 1B. The study further demonstrated the convenience of using the C. elegans-C. albicans infection model to screen C. albicans mutants for genes implicated in virulence. The screening identified the role of two novel genes; ADA2 and CAS5 in Candida virulence7. Interestingly, an ADA2 deletion mutant showed normal filamentation in vitro but showed attenuated filamentation inside C. elegans, highlighting the utility of C. elegans as a substitute in vivo host for characterisation of C. albicans virulence7. Other Candida virulence factors have also been identified using C. elegans, including the co-transcription factor known as Mediator Med31 and the mitochondrial outer membrane SAM (Sorting and Assembly Machinery) complex subunit (Sam37), which were both shown to have an impact on C. albicans virulence16,17. The Sam37 results were also confirmed using a mammalian infection model.
|
Our group has also extended the use of C. elegans to study polymicrobial infections18. Our results have revealed interesting interactions between two diverse and clinically important organisms, the bacterium Acinetobacter baumannii and the fungus C. albicans. We showed that A. baumannii inhibits the ability of Candida to form filaments in C. elegans. This attenuates the virulence of Candida as determined by reduced C. elegans killing18.
C. elegans is a powerful model system to study host pathogen interactions and has excellent predictive value for microbial pathogenesis in mammalian models. It is widely used for illustrating mechanisms of virulence in diverse pathogenic organisms including the human fungal pathogen C. albicans, enabling the identification of novel mechanisms that would not necessarily be determined using in vitro assays.
References
[1] Leroy, O. et al. (2009) Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006). Crit. Care Med. 37, 1612–1618.| Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006).Crossref | GoogleScholarGoogle Scholar | 19325476PubMed |
[2] Calderone, R.A. (2002) Candida and candidiasis. 2002, Washington, DC: ASM Press.
[3] Berman, J. and Sudbery, P.E. (2002) Candida albicans: A molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3, 918–932.
| Candida albicans: A molecular revolution built on lessons from budding yeast.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XptFCjt74%3D&md5=6efa06db53249f8c9e8b79ff00881758CAS | 12459722PubMed |
[4] Larriba, G. et al. (2000) Candida albicans molecular biology reaches its maturity. Int. Microbiol. 3, 247–252.
| 1:CAS:528:DC%2BD3MXks1Kgtb4%3D&md5=801e85f85ddcab96e41d8d48389bed98CAS | 11334309PubMed |
[5] Dalle, F. et al. (2010) Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 12, 248–271.
| Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFGlu7s%3D&md5=0b46dc69a2b4f1488cdb480c9ddef635CAS | 19863559PubMed |
[6] Gow, N.A.R. et al. (2002) Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5, 366–371.
| Fungal morphogenesis and host invasion.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XlvVequr0%3D&md5=241abc7f6f98516ca5c78d1ae7f678b2CAS |
[7] Pukkila-Worley, R. et al. (2009) Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot. Cell 8, 1750–1758.
| Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFSku73K&md5=e9197dfe8f15bdb93a9d5c2f235ecbadCAS | 19666778PubMed |
[8] Sifri, C.D. et al. (2005) The worm has turned-microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13, 119–127.
| The worm has turned-microbial virulence modeled in Caenorhabditis elegans.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhslehsro%3D&md5=ea76649dcd19b6569560660208a5f014CAS | 15737730PubMed |
[9] Fuchs, B.B. and Mylonakis, E. (2006) Using non-mammalian hosts to study fungal virulence and host defense. Curr. Opin. Microbiol. 9, 346–351.
| Using non-mammalian hosts to study fungal virulence and host defense.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xns1Kju70%3D&md5=40b591428b661bde9a9c9e6c7695ce8eCAS | 16814595PubMed |
[10] Aballay, A. and Ausubel, F.M. (2002) Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr. Opin. Microbiol. 5, 97–101.
| Caenorhabditis elegans as a host for the study of host-pathogen interactions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XhtVKqsbk%3D&md5=e8b53d199a1d4bde6496110ed9fbf3e7CAS | 11834377PubMed |
[11] Ewbank, J.J. and Zugasti, O. (2011) C. elegans: model host and tool for antimicrobial drug discovery. Dis. Model. Mech. 4, 300–304.
| C. elegans: model host and tool for antimicrobial drug discovery.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXntlKmsrg%3D&md5=d6c0001c7858e0b118600769c84dfb56CAS | 21504910PubMed |
[12] Mylonakis, E. et al. (2002) Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99, 15675–15680.
| Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjvFSi&md5=76af29fda001ccf0aa3ea993dd715225CAS | 12438649PubMed |
[13] Johnson, C.H. et al. (2009) Histoplasma capsulatum and Caenorhabditis elegans: a simple nematode model for an innate immune response to fungal infection. Med. Mycol. 47, 808–813.
| Histoplasma capsulatum and Caenorhabditis elegans: a simple nematode model for an innate immune response to fungal infection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhs1aqt7jJ&md5=f960c4bf31bda5b8ce1ae2de3fbe04c0CAS | 20028234PubMed |
[14] Huang, X. et al. (2014) Caenorhabditis elegans: a simple nematode infection model for Penicillium marneffei. PLoS ONE 9, e108764.
| Caenorhabditis elegans: a simple nematode infection model for Penicillium marneffei.Crossref | GoogleScholarGoogle Scholar | 25268236PubMed |
[15] Breger, J. et al. (2007) Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3, e18.
| Antifungal chemical compounds identified using a C. elegans pathogenicity assay.Crossref | GoogleScholarGoogle Scholar | 17274686PubMed |
[16] Uwamahoro, N. et al. (2012) The functions of mediator in Candida albicans support a role in shaping species-specific gene expression. PLoS Genet. 8, e1002613.
| The functions of mediator in Candida albicans support a role in shaping species-specific gene expression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XlslejtLw%3D&md5=212104b929c99d3fd6a815a52143a1cfCAS | 22496666PubMed |
[17] Qu, Y. et al. (2012) Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence. Eukaryot. Cell 11, 532–544.
| Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xlslams74%3D&md5=0d808bb1b9d4936427c178cdf422f3f8CAS | 22286093PubMed |
[18] Peleg, A.Y. et al. (2008) Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105, 14585–14590.
| Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXht1Sgt7%2FN&md5=8163b2e889b35eb88393d62593fc77e8CAS | 18794525PubMed |
Biographies
Farkad Bantun is a PhD student in the Peleg Lab at Monash University, Melbourne, and is working on the genetic mechanisms driving the morphological transition of Candida from yeast to a filamentous form using Caenorhabditis elegans as an in vivo model. He finished his schooling in Saudi Arabia where he also completed his bachelor degree in Medical Technology. He was awarded a scholarship to complete his postgraduate studies at Monash University. He also finished a Masters degree in Laboratory Medicine at RMIT University, Melbourne.
Sanjiveeni Dhamgaye is a postdoctoral research fellow in the Peleg lab working on polymicrobial biofilms and Candida pathogenesis. She was born in New Delhi, India, and after her schooling, pursued a career in Science. She completed her PhD at Jawaharlal Nehru University, New Delhi, India, with a focus on ‘Regulation of Multidrug resistance in yeast’ with Prof. Rajendra Prasad.
Professor Anton Y Peleg is the Director of the Department of Infectious Diseases at the Alfred Hospital and Monash University, and is a research group leader in the Department of Microbiology, Monash University. His research spans clinical to basic research, with a focus on hospital-acquired infections, antimicrobial resistance, infections in immunocompromised hosts and understanding mechanisms of disease caused by hospital pathogens, including Candida, Staphylococcus aureus and Acinetobacter baumannii. His group has expertise in the use of non-mammalian model systems to study microbial pathogenesis.


