Next generation sequencing in single cell parasite disease investigations
Jan ŠlapetaParasitology Laboratory
McMaster Building B14
Faculty of Veterinary Science
University of Sydney
NSW 2006, Australia
Email: jan.slapeta@sydney.edu.au
Microbiology Australia 34(4) 192-193 https://doi.org/10.1071/MA13067
Published: 18 September 2013
Single cell parasites, also referred as protozoa, are ubiquitous. They are parasites of animals and humans, causing significant disease such as malaria and toxoplasmosis1. In farm animal medicine and human medicine, specific diagnostic tests have been developed to detect many of these diseases. Unfortunately, the role of protozoal agents in wildlife disease is poorly understood and diagnosis is confounded by the lack of basic knowledge of parasite distribution and morphological identification2,3. Therefore we are pursuing a new approach using 'state of the art' next generation sequencing to address existing limitations. The approach relies on microbial community DNA analysis as a faster and more economical method compared with development of traditional species-specific diagnostic methodologies.
Protozoa comprise diverse lineages of eukaryotes, but their morphology under a light microscope might be ambiguous because of their relatively small size and featureless characteristics. In this setting, emerging protozoal disease relies on DNA identification. The only weakness is that the approach is underpinned by a requirement that reference databases are sufficiently complete. The remaining challenge of obtaining, amplifying and sequencing the DNA requires some thoughtful consideration. Traditionally, characterisation of protozoa in wildlife relies on an initial speculation about the group of protozoa present in the sample, i.e. blood smear, histological section or organ biopsy. This preliminary speculation is used to apply existing PCR primer combinations to target only the conservative DNA region of the protozoan parasite. For a parasite like Toxoplasmagondii present in humans and animals, this task is quite straightforward4. On the other hand, there are many more protozoa in wildlife and other animals for which no such groundwork and published methods are available. A guessing game of ‘trial and error’ PCR begins and too often ends with unsatisfactory results. This phase is time consuming and for wildlife investigation often too costly.
An emerging alternative solution now exists. Rather than amplification of the parasite DNA only, we can use generic primers that amplify a conservative region of all eukaryotes in the specimens5. Theoretically, such generic primers amplify both host and parasites. To overcome the overwhelming presence of the host DNA amplification compared with the relatively sparse parasite we need technology that will allow us to sequence as much of the amplified DNA as possible. Previously prohibitively expensive, rapidly falling costs have made next generation sequencing (NGS) technologies for sequencing of community DNA accessible even to poorly funded wildlife investigations. Using NGS we randomly sequence lots of individual threads of the DNA within the sample. The user can select how many such DNA threads should be sequenced. For example, if 3,000 threads are sequences (at a cost of 5c per DNA thread) from a single DNA sample from a sample with an ‘unknown’ protozoan, we have a good chance of detecting protozoan DNA despite the plethora of host DNA present, as long as it is there in the ratio 1:3000. Currently, this approach utilising 454 amplicon pyrosequencing based on small subunit rDNA is being trialed in the Parasitology Laboratory at the Faculty of Veterinary Science with very promising results. Recently, we used this approach to document the presence of an unusual protozoan parasite infecting coral on the Great Barrier Reef; these parasites were previously only known from the Caribbean coral6. Projects using this approach now include a quest to document unidentified protozoa of some iconic Australian wildlife species such as the platypus and echidna.
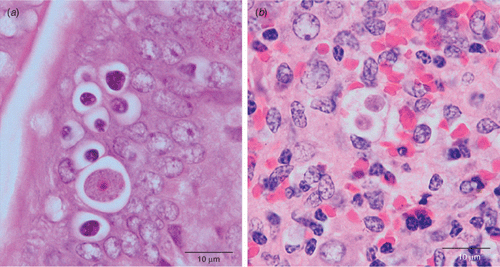
In collaboration with Larry Vogelnest at Taronga Zoo and Richard Whittington at The University of Sydney, we are currently undertaking an investigation into an enigmatic but fatal disease in echidnas. The disease syndrome is recognised by clinicians and pathologists as systemic coccidiosis, with presence of coccidian-like protozoa in multiple organs detectable by histopathology7. Coccidian parasites are protozoa in the phylum Apicomplexa. Echidnas are known to have at least two distinct coccidia commonly infect their gastrointestinal tract8 (Fig. 1). Identity of the extraintestinal stages compared with the intestinal coccidia is currently being investigated using an NGS approach utilising 454 amplicon pyrosequencing. Our material is processed through the Diversity Profiling Service at AGRF (Australian Genome Research Facility Ltd) and sequence data analysed using the Faculty’s clustering pipeline. Outcomes of this NGS approach allow us to match the blueprints of intestinal coccidia with those in host tissues as well as capture other non-coccidian parasites historically associated with this syndrome. Overall, this proof of concept study has already outperformed a traditional approach utilising coccidian specific primers and PCR conducted in parallel (S Severimuttu, C Sangster, K Rose, L Vogelnest, R Whittington, J Šlapeta, unpublished data).
|
The approach is an adaptation of the microbial community assessment that greatly benefited from cloning-independent and massively parallel approach, facilitated by 454 pyrosequencing technology9. The 454 amplicon sequencing technology based on hypervariable regions of the rDNA has been successfully applied in investigative ecology of marine single cell eukaryotes and picoeukaryotes10–12 and now the cost of the technology allows its application in disease investigation. In fact, amplicon sequencing using 454 pyrosequencing is demonstrated to be superior in detecting rare species than any other community-based technique. Therefore, it has the attributes necessary to be useful in wildlife disease investigation where the samples, host and pathogens are diverse.
Baseline data, such as parasite diversity, are fundamental in disease diagnosis and understanding the functional microbial associations in the integrated investigation of interactions between host, pathogen and environment. The marriage of the emerging technologies with wildlife disease investigation has the opportunity to advance previously described wildlife syndromes whose causality is yet to be documented.
References
[1] Adlard, R.D. and O’Donoghue, P.J. (1998) Perspectives on the biodiversity of parasitic protozoa in Australia. Int. J. Parasitol. 28, 887–897.| Perspectives on the biodiversity of parasitic protozoa in Australia.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK1czjvVKmtA%3D%3D&md5=eae583d407794086a5e996f11b8c51d1CAS | 9673868PubMed |
[2] Zhu, B.Y. et al. (2009) Looks can deceive: molecular identity of an intraerythrocytic apicomplexan parasite in Australian gliders. Vet. Parasitol. 159, 105–111.
| Looks can deceive: molecular identity of an intraerythrocytic apicomplexan parasite in Australian gliders.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnsVSquw%3D%3D&md5=c3a998bf1453dc1cd1c9ef1152d8c832CAS | 19028015PubMed |
[3] Hartigan, A. et al. (2012) Myxozoan parasite in brain of critically endangered frog. Emerg. Infect. Dis. 18, 693–695.
| Myxozoan parasite in brain of critically endangered frog.Crossref | GoogleScholarGoogle Scholar | 22469079PubMed |
[4] Miller, M.A. et al. (2008) Type X Toxoplasma gondii in a wild mussel and terrestrial carnivores from coastal California: new linkages between terrestrial mammals, runoff and toxoplasmosis of sea otters. Int. J. Parasitol. 38, 1319–1328.
| Type X Toxoplasma gondii in a wild mussel and terrestrial carnivores from coastal California: new linkages between terrestrial mammals, runoff and toxoplasmosis of sea otters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXps12isrw%3D&md5=ff190da679ef736fc864d62cc2e6195cCAS | 18452923PubMed |
[5] Sun, Y. et al. (2011) Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol. Biol. 733, 129–141.
| Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXntVChur8%3D&md5=7edacd60f642a568a36e8b34eafade08CAS | 21431767PubMed |
[6] Šlapeta, J. and Linares, M. (2013) Combined amplicon pyrosequencing assays reveal presence of the apicomplexan “type-N” (cf. Gemmocystis cylindrus) and Chromera velia on the Great Barrier Reef, Australia. PLoS ONE , .
| Combined amplicon pyrosequencing assays reveal presence of the apicomplexan “type-N” (cf. Gemmocystis cylindrus) and Chromera velia on the Great Barrier Reef, Australia.Crossref | GoogleScholarGoogle Scholar |
[7] Vogelnest, L. and Woods, R., eds (2008) Medicine of Australian Mammals, CSIRO Publishing.
[8] Debenham, J.J. et al. (2012) Year-long presence of Eimeria echidnae and absence of Eimeria tachyglossi in captive short-beaked echidnas (Tachyglossus aculeatus). J. Parasitol. 98, 543–549.
| Year-long presence of Eimeria echidnae and absence of Eimeria tachyglossi in captive short-beaked echidnas (Tachyglossus aculeatus).Crossref | GoogleScholarGoogle Scholar | 22236183PubMed |
[9] Margulies, M. et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380.
| 1:CAS:528:DC%2BD2MXpvFOrt7s%3D&md5=dea90f3c1f833bc33f050a7be0550989CAS | 16056220PubMed |
[10] Amaral-Zettler, L.A. et al. (2011) Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J. 5, 42–50.
| Microbial community structure across the tree of life in the extreme Rio Tinto.Crossref | GoogleScholarGoogle Scholar | 20631808PubMed |
[11] Cheung, M.K. et al. (2010) Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 4, 1053–1059.
| Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing.Crossref | GoogleScholarGoogle Scholar | 20336159PubMed |
[12] Stoeck, T. et al. (2009) Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 7, 72.
| Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities.Crossref | GoogleScholarGoogle Scholar | 19886985PubMed |
Biography
Jan Šlapeta joined the Parasitology team in the Faculty of Veterinary science at The University of Sydney in 2007. He has a broad understanding of the biology of parasites of both medical and veterinary importance, as well as the diseases caused by them. He has experience in several research laboratories, including the NIH in the USA, the CNRS in France and the University of Technology in Sydney. Jan specialises in the molecular identification and evolution of protozoan parasites. His diagnostic techniques and biodiversity studies have received worldwide interest. He has a particular interest in applications of molecular biology towards elucidating the unique properties of parasites of medical and veterinary importance.


