An updated view on bacterial glycogen structure
Liang Wang A B C F and Michael J Wise D EA Department of Bioinformatics, School of Medical Informatics and Engineering, Xuzhou Medical University, Xuzhou, Jiangsu 221000, China
B Jiangsu Key Laboratory of New Drug Research and Clinical Pharmacy, School of Pharmacy, Xuzhou Medical University, Xuzhou, Jiangsu 221000, China
C Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi, Jiangsu 214122, China
D The Marshall Centre for Infectious Diseases Research and Training, University of Western Australia, Perth, WA 6009, Australia
E Computer Science and Software Engineering, Faculty of Engineering and Mathematical Sciences, University of Western Australia, Perth, WA 6009, Australia
F Email: leonwang@xzhmu.edu.cn
Microbiology Australia 40(4) 195-199 https://doi.org/10.1071/MA19056
Published: 8 November 2019
Glycogen is a homogenous and multi-disperse polysaccharide that is present in many clinically significant bacteria, such as Escherichia coli, Vibrio cholera and Mycobacterium tuberculosis. Its structure and metabolism have been linked with environmental viability, intracellular growth, pathogenicity and transmission capacity. However, due to the harsh extraction conditions and also the inconsistent methods for structure characterisation, understanding of bacterial glycogen structure and its association with bacterial metabolism and physiology has been hindered. Here we gave a concise overview of bacterial glycogen structure with a focus on its recently discovered higher level organisation, α particle. Standardised procedures for glycogen extraction and structure detection are also highlighted.
Glycogen metabolism
Glycogen is a widespread homogeneous polysaccharide and plays important roles in bacterial energy reserves and carbon supply with little effect on cellular osmolarity1. Experimental studies have linked glycogen to bacterial environmental survival, intracellular growth, pathogenicity, transmission, etc., although controversial observations exist2. Since glycogen plays a central role in bacterial carbohydrate metabolism, its synthesis and degradation are highly regulated. A systematic analysis of 1202 bacterial species proteomes confirmed that 402 of them harbour a complete set of enzymes in the classical glycogen metabolism pathway3. This pathway involves five enzymes operating concurrently to promote glycogen synthesis: glucose-1-phosphate adenylyltransferase (GlgC), glycogen synthase (GlgA), glycogen branching enzyme (GlgB); and degradation: glycogen phosphorylase (GlgP), and glycogen debranching enzyme (GlgX)1. A second non-classical and widespread glycogen synthesis pathway was unexpectedly found in Mycobacterium tuberculosis, which involves trehalose synthase (TreS), maltokinase (Mak1 or Pep2) and maltosyltransferase (GlgE)4. This pathway established a solid connection among glucose, trehalose, maltose and glycogen metabolism. Another branched α-glucan pathway involves two paralogues, glycogen synthase (Rv3032) and glycogen branching enzyme (Rv3031), which generates methyl-glucose lipopolysaccharide and is associated with fatty acid metabolism (Figure 1)4,5. However, its connection with cytosolic glycogen metabolism is still under investigation. A comprehensive genome-wide screening of genes affecting glycogen accumulation, based on 3985 single-gene knockout mutants of nonessential genes in E. coli K-12 (Keio Collection), confirmed that 35 genes were related with glycogen-excess phenotypes while 30 genes were related with glycogen-deficient phenotypes6. Of the 65 gene products, their functions are mainly involved in direct glycogen synthesis and/or degradation, energy production, amino acid provision and cell envelope integrity6. Another systematic study using E. coli gene expression library ASKA observed that upregulation of 86 genes could influence glycogen accumulation7. The genes fell into the functional categories of general stress and stringent responses, aggregative and social behaviour, and intracellular communication, etc.7. Thus, glycogen metabolism is a highly interconnected process with a wide variety of cellular interactions. Among those sophisticated interactions, glycogen breakdown plays an important role in the interactions between the host and pathogenic bacteria, though the mechanisms and roles of glycogen during infection are still not fully elucidated1.

|
Glycogen structure
Glycogen is characterised by a hyperbranched structure with α-1,4-glycosidic linkages at the linear chains and α-1,6-glycosidic linkages at the branching points1. As an efficient energy storage molecule, a good structure-function relationship should be achieved for optimally providing energy and carbon source8. Although multiple models have been proposed to describe glycogen structure, such as the classical Whelan model, fractal structure model, and the newly proposed Monte-Carlo simulation, there is no settled model that accurately describes the polydisperse structures of glycogen, including particle size and chain length distributions, etc.3,9. Currently, three structural levels have been defined in glycogen found in the liver of higher organisms, which are (1) linear α-D-glucosyl chains through α-1,4-glycosidic bonds, (2) highly branched β particles of around 20 nm in diameter, and (3) β-particle-aggregated rosette-shaped α particles up to 300 nm in diameter10. However, the molecular basis for the aggregation of β particles into α particles is still under investigation. Glycogen particles are also associated with a group of proteins (known as the glycogen proteome) that regulate its biological functions11. Proteomic analysis of the rat, mouse and human liver inferred that a self-glycosylating homodimer protein, glycogenin, is the binding agent on the surface of β particles to form α particles12. Intriguingly, no coding genes for glycogenin or its homologs have ever been identified in bacterial genomes; it is glycogen synthase that is involved in glucan initiation and elongation in bacteria13.
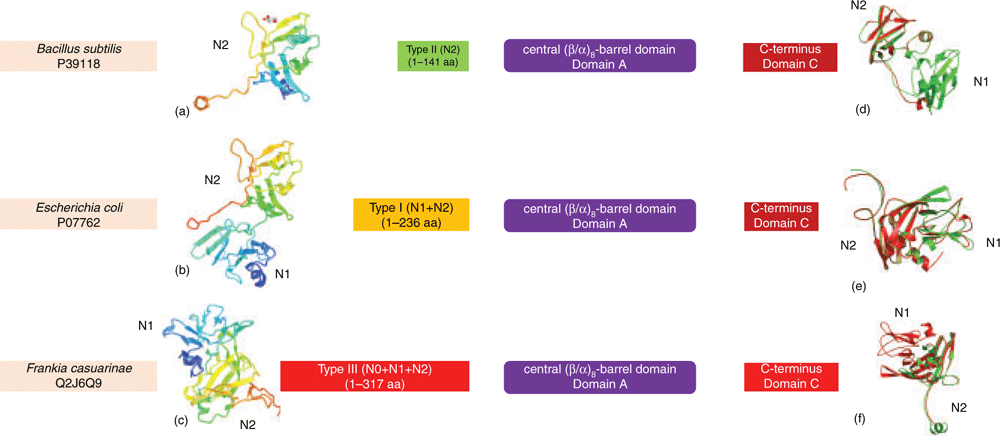
Until recently, only β particles were reported in bacteria, containing up to 55 000 glucosyl units with molecular mass of around 107–108 Daltons and varying in diameter from 20 to 50 nm1. The average length of linear chains, gc, in β particles varies between 6.6 to 23.5 glucose units in a species-dependent manner3. Correlation analysis between gc and bacterial environmental viability showed that bacteria accumulating glycogen with short gc have greater survival times in the external environment3. It was argued that short gc glycogen could lead to the formation of more compact β particles with higher branching degrees3,14. These properties facilitate the slow utilisation of glycogen by hindering its degradation14, enhancing bacterial survivability3. Although maintaining glycogen structure requires a coordinated action of enzymatic activities and different structural proteins, GlgB plays a dominant role in chain length distribution patterns and corresponding branching degrees15. Recent analysis showed that domain organisation of GlgB is conserved, which consists of an N-terminus, a central (β/α)-barrel region and a C-terminus15. Based on N-terminal length variation, two groups of glycogen branching enzymes (GBEs) were initially identified: Group 1 with duplicated CBM48 domains (N1 and N2 domains) and Group 2 with a single CBM48 domain (N2 domains)16. A longer GBE N-terminus was further found to contribute to the transfer of shorter side chains, leading to shorter gc glycogen17. A large comparison of 9387 bacterial GlgBs revealed that there might exist a third group with a longer N-terminus (N0, N1, and N2 domains) than that in Group 1 GlgBs (Figure 2)15. Thus, N-terminal diversity could be one of the determinant factors for the polydispersity of glycogen structures leading to the heterogeneity of degradation rates. However, experimental evidence is needed to validate the hypothesis.
|
Glycogen extraction methods
A variety of extraction methods have been used for bacterial glycogen isolation, such as the harsh boiling method with 30% potassium hydroxide solution (KOH-HW)18 and the comparatively mild trichloroacetic acid extraction methods with cold or hot water (TCA-CW or TCA-HW)19. Less commonly used reagents such as sodium dodecyl sulfate (SDS), thymol or glycine for bacterial glycogen extraction have also be used in sporadic studies20–22. Harsh extraction procedures with alkali, acid, and/or heat can degrade glycogen primary and tertiary structures23, which could compromise experimental results and conclusions. On the other hand, the use of diverse extraction methods makes it difficult to compare glycogen structures due to the lack of uniform procedures. This is particularly the case with the fragile glycogen-associated proteins if one aims to investigate the functions of glycogen-associated proteomes. In higher organisms, sucrose density gradient ultracentrifugation with cold water (SDGU-CW) has been confirmed as less- or non-degradative method and is regularly used for glycogen extraction with the purpose of α particle study or glycogen proteomic analysis23. SDGU-CW was previously used for glycogen extraction in Selenomonas ruminantium and Fibrobacter succinogenes20,24. Interestingly, glycogen morphology via transmission electron microscopy (TEM) in both studies showed the possible existence of rosette-shaped α particles in bacteria, although this phenomenon did not garner much attention20,24. Our lab recently explored this issue by comparing different extraction methods and confirmed the existence of bacterial glycogen α particles extracted via the SDGU-CW method in E. coli (Figure 3)9. Thus, we suggested that the mild SDGU-CW extraction method should be adopted as a standard method in bacterial glycogen study so as to reveal genuine comparative structural features. In addition, the discovery of glycogen α particles in both prokaryotes and eukaryotes indicates that any organism needing to store and then release glucose might have similar α and β particle structures: a type of convergent evolution, regardless of the presence of glycogenin9. Since α particles have been linked with release rate of glucose during degradation stage, further exploration of the formation mechanisms of glycogen α particles in bacteria would be essential for better understanding its functions in bacterial physiology and pathogenicity. Relevant questions to ask include whether under certain circumstances more α particles will be synthesised while β particles will be more preferred in other situations based on the regulation of the activities and contents of α-particle forming enzyme(s).

|
Glycogen structural characterisation
Glycogen structure can be measured from different aspects, such as morphology, branching degree (percentage of α-1, 6-glycosidic linkages), branch length distribution, and particle size distribution, etc. Glycogen morphology, such as α and β particles, can be most easily characterised using TEM9. Distributions of glycogen sizes can also be generated via histograms based on TEM images. However, with the latter method it is rather difficult to achieve quantitative results and it cannot give genuine particle sizes. Periodate oxidation is a commonly used method in the structural study of non-ionic polysaccharides in carbohydrate chemistry. The methods are done using sodium metaperiodate as the oxidising agent, and the presence of α-1, 6-glycosidic linkages can be confirmed by the free hydroxyl groups resulting from the consumption of periodate ions during the periodate oxidation reaction25. From this, glycogen branching degree can be calculated. In addition, methylation26, reducing end assay19, and nuclear magnetic resonance (NMR)27 can also be used for measuring glycogen branching degree. In terms of chain lengths, iodine staining was originally used with the aid of spectrophotometry. It was shown that polysaccharide chains consisting of 8 to 12 glucose units stain a reddish colour at the wavelength of 520 nm, while at a length of 30 to 35 units, the stain appears blue, with a peak at 600 nm, similar to amylose28. Although multiple studies have used this method to infer glycogen branching features, the method has mainly been applied to the analysis of amylose and amylopectin due to the weak interaction of glycogen with iodine29. On the other hand, fluorophore-assisted capillary electrophoresis (FACE) is an advanced method for accurately quantifying the chain-length distributions of isoamylase-treated glycogen particles19. The average chain length of glycogen can be calculated based on percentages of oligosaccharides with specific degrees of polymerisation generated via FACE3. Until recently, FACE has been the only method that can give an accurate description of glycogen primary structure.
Size exclusion chromatography (SEC), together with a differential refractive index (DRI) detector, separates glycogen solely on hydrodynamic volumes Vh or the corresponding radius Rh30. With a multiple-angle laser light scattering (MALLS) detector, weight-average molecular weight (Mw) can be measured and molecular density of glycogen particles calculated as ρ(Rh) = Mw(Rh) × 4/3πRh3. Thus, this method gives a quantitative overview of glycogen particle size distribution and should be preferred for glycogen analysis. The method can also be used for detecting fragility and stability of glycogen α particles. That is, fragile α particles are easily degraded into β particles after treatment with hydrogen bond disruptors like dimethyl sulfoxide (DMSO), which is reflected in the shift of glycogen particles toward smaller sizes on SEC graphs10. Our recent study also shows that both fragile and stable α particles exist in E. coli with unclear functions (Figure 4)9.

|
Summary and future perspectives
Glycogen is a central energy reserve in bacteria. Understanding the interactions between glycogen structure and metabolism has apparent clinical significance due to its associations with viability and virulence of bacterial pathogens. Lack of standardised extraction procedures and detection methods hinders the comprehensive understanding of glycogen functions across bacterial species. The pros and cons of various glycogen extraction procedures and detection methods were compared in recent studies and the SDGU-CW method is recommended for glycogen isolation due to its comparatively mild extraction conditions. On the other hand, SEC is suggested for measuring size, weight, and density of glycogen particles. In addition, TEM is recommended for studying glycogen morphology, while FACE is most suitable for dissecting glycogen primary structure. Most importantly, the discovery of fragile and stable glycogen α particles in bacteria creates a new avenue in the bacterial glycogen research field, from where it would be interesting for us to investigate how glucose concentration in the culture influences bacterial glycogen α particles formation, how glycogen α particles change in different stages of bacterial life, and what proteins are responsible for the formation and fragility of α particles in bacteria, etc. These studies might lead us to a better understanding of how glycogen contributes to bacterial environmental survival, intracellular growth, pathogenicity and transmission.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Excellent Researcher’s Startup Foundation at Xuzhou Medical University (No. D2016007), Natural Science Foundation of Jiangsu Province (No. BK20180997), the Innovative and Entrepreneurial Talent Scheme in Jiangsu Province (2017), National Natural Science Foundation of China (No. 31900022), and the Open Project from the Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education at Jiangnan University (No. KLCCB-KF201902).
References
[1] Park, K.H. (2015) Roles of enzymes in glycogen metabolism and degradation in Escherichia coli. J. Appl. Glycosci. 62, 37–45.| Roles of enzymes in glycogen metabolism and degradation in Escherichia coli.Crossref | GoogleScholarGoogle Scholar |
[2] Wilson, W.A. et al. (2010) Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 34, 952–985.
| Regulation of glycogen metabolism in yeast and bacteria.Crossref | GoogleScholarGoogle Scholar | 20412306PubMed |
[3] Wang, L. and Wise, M.J. (2011) Glycogen with short average chain length enhances bacterial durability. Naturwissenschaften 98, 719–729.
| Glycogen with short average chain length enhances bacterial durability.Crossref | GoogleScholarGoogle Scholar | 21808975PubMed |
[4] Chandra, G. et al. (2011) Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology 157, 1565–1572.
| Unexpected and widespread connections between bacterial glycogen and trehalose metabolism.Crossref | GoogleScholarGoogle Scholar | 21474533PubMed |
[5] Wang, L. et al. (2019) Bioinformatics analysis of metabolism pathways of archaeal energy reserves. Sci. Rep. 9, 1034.
| Bioinformatics analysis of metabolism pathways of archaeal energy reserves.Crossref | GoogleScholarGoogle Scholar | 30705313PubMed |
[6] Eydallin, G. et al. (2007) Genome-wide screening of genes affecting glycogen metabolism in Escherichia coli K-12. FEBS Lett. 581, 2947–2953.
| Genome-wide screening of genes affecting glycogen metabolism in Escherichia coli K-12.Crossref | GoogleScholarGoogle Scholar | 17543954PubMed |
[7] Eydallin, G. et al. (2010) Genome-wide screening of genes whose enhanced expression affects glycogen accumulation in Escherichia coli. DNA Res. 17, 61–71.
| Genome-wide screening of genes whose enhanced expression affects glycogen accumulation in Escherichia coli.Crossref | GoogleScholarGoogle Scholar | 20118147PubMed |
[8] Meléndez, R. et al. (1998) Physical constraints in the synthesis of glycogen that influence its structural homogeneity: a two-dimensional approach. Biophys. J. 75, 106–114.
| Physical constraints in the synthesis of glycogen that influence its structural homogeneity: a two-dimensional approach.Crossref | GoogleScholarGoogle Scholar | 9649371PubMed |
[9] Wang, L. et al. (2019) Molecular structure of glycogen in Escherichia coli. Biomacromolecules 20, 2821–2829.
| Molecular structure of glycogen in Escherichia coli.Crossref | GoogleScholarGoogle Scholar | 31244022PubMed |
[10] Hu, Z. et al. (2018) Diurnal changes of glycogen molecular structure in healthy and diabetic mice. Carbohydr. Polym. 185, 145–152.
| Diurnal changes of glycogen molecular structure in healthy and diabetic mice.Crossref | GoogleScholarGoogle Scholar | 29421051PubMed |
[11] Stapleton, D. et al. (2010) Analysis of hepatic glycogen-associated proteins. Proteomics 10, 2320–2329.
| Analysis of hepatic glycogen-associated proteins.Crossref | GoogleScholarGoogle Scholar | 20391537PubMed |
[12] Tan, X. et al. (2018) Proteomic investigation of the binding agent between liver glycogen β particles. ACS Omega 3, 3640–3645.
| Proteomic investigation of the binding agent between liver glycogen β particles.Crossref | GoogleScholarGoogle Scholar | 30023874PubMed |
[13] Ugalde, J.E. et al. (2003) De novo synthesis of bacterial glycogen: Agrobacterium tumefaciens glycogen synthase is involved in glucan initiation and elongation. Proc. Natl. Acad. Sci. USA 100, 10659–10663.
| De novo synthesis of bacterial glycogen: Agrobacterium tumefaciens glycogen synthase is involved in glucan initiation and elongation.Crossref | GoogleScholarGoogle Scholar | 12960388PubMed |
[14] Martinez-Garcia, M. et al. (2017) The glycogen of Galdieria sulphuraria as alternative to starch for the production of slowly digestible and resistant glucose polymers. Carbohydr. Polym. 169, 75–82.
| The glycogen of Galdieria sulphuraria as alternative to starch for the production of slowly digestible and resistant glucose polymers.Crossref | GoogleScholarGoogle Scholar | 28504180PubMed |
[15] Wang, L. et al. (2019) Structure and evolution of glycogen branching enzyme N-termini from bacteria. Front. Microbiol. 9, 3354.
| Structure and evolution of glycogen branching enzyme N-termini from bacteria.Crossref | GoogleScholarGoogle Scholar | 30692986PubMed |
[16] Leggio, L.L. et al. (2002) A structural model for the N-terminal N1 module of E. coli glycogen branching enzyme. Biologia 57, 10.
[17] Devillers, C.H. et al. (2003) Characterization of the branching patterns of glycogen branching enzyme truncated on the N-terminus. Arch. Biochem. Biophys. 418, 34–38.
| Characterization of the branching patterns of glycogen branching enzyme truncated on the N-terminus.Crossref | GoogleScholarGoogle Scholar | 13679080PubMed |
[18] Lou, J. et al. (1997) Glycogen formation by the ruminal bacterium Prevotella ruminicola. Appl. Environ. Microbiol. 63, 1483–1488.
| 16535575PubMed |
[19] Wang, L. et al. (2015) Influence of in situ progressive N-terminal is still controversial truncation of glycogen branching enzyme in Escherichia coli DH5α on glycogen structure, accumulation, and bacterial viability. BMC Microbiol. 15, 96.
| Influence of in situ progressive N-terminal is still controversial truncation of glycogen branching enzyme in Escherichia coli DH5α on glycogen structure, accumulation, and bacterial viability.Crossref | GoogleScholarGoogle Scholar | 25947105PubMed |
[20] Kamio, Y. et al. (1981) Structure of glycogen produced by Selenomonas ruminantium. Agric. Biol. Chem. 45, 209–216.
| Structure of glycogen produced by Selenomonas ruminantium.Crossref | GoogleScholarGoogle Scholar |
[21] Sambou, T. et al. (2008) Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Mol. Microbiol. 70, 762–774.
| Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice.Crossref | GoogleScholarGoogle Scholar | 18808383PubMed |
[22] Whyte, J.N.C. and Strasdine, G.A. (1972) An intracellular α-D-glucan from Clostridium botulinum, type E. Carbohydr. Res. 25, 435–441.
| An intracellular α-D-glucan from Clostridium botulinum, type E.Crossref | GoogleScholarGoogle Scholar |
[23] Tan, X. et al. (2016) A new non-degradative method to purify glycogen. Carbohydr. Polym. 147, 165–170.
| A new non-degradative method to purify glycogen.Crossref | GoogleScholarGoogle Scholar | 27178921PubMed |
[24] Gong, J. and Forsberg, C.W. (1993) Separation of outer and cytoplasmic membranes of Fibrobacter succinogenes and membrane and glycogen granule locations of glycanases and cellobiase. J. Bacteriol. 175, 6810–6821.
| Separation of outer and cytoplasmic membranes of Fibrobacter succinogenes and membrane and glycogen granule locations of glycanases and cellobiase.Crossref | GoogleScholarGoogle Scholar | 8226622PubMed |
[25] Northcote, D.H. (1953) The molecular structure and shape of yeast glycogen. Biochem. J. 53, 348–352.
| The molecular structure and shape of yeast glycogen.Crossref | GoogleScholarGoogle Scholar | 13032077PubMed |
[26] Manners, D.J. (1989) Some aspects of the structure of starch and glycogen. J. Jpn. Soc. Starch Sci. 36, 311–323.
| Some aspects of the structure of starch and glycogen.Crossref | GoogleScholarGoogle Scholar |
[27] Sillerud, L.O. and Shulman, R.G. (1983) Structure and metabolism of mammalian liver glycogen monitored by carbon-13 nuclear magnetic resonance. Biochemistry 22, 1087–1094.
| Structure and metabolism of mammalian liver glycogen monitored by carbon-13 nuclear magnetic resonance.Crossref | GoogleScholarGoogle Scholar | 6838841PubMed |
[28] Swanson, M.A. (1948) Studies on the structure of polysaccharides: IV. Relation of the iodine color to the structure. J. Biol. Chem. 172, 825–837.
| 18901205PubMed |
[29] Craig, S.A.S. et al. (1988) The iodine-staining properties and fine structure of some mammalian and invertebrate glycogens. Carbohydr. Res. 179, 327–340.
| The iodine-staining properties and fine structure of some mammalian and invertebrate glycogens.Crossref | GoogleScholarGoogle Scholar |
Biographies
Associate Professor Liang Wang graduated with a PhD degree from the University of Western Australia in 2014, under the supervision of Associate Professor Michael J Wise and Associate Professor Charlene M Kahler. He worked as a postdoctoral researcher at Concordia University (Montreal, Canada) and then a research associate at Curtin University (Perth, Australia). He is currently the director of the Department of Bioinformatics at Xuzhou Medical University, China. He is also an adjunct researcher at Jiangsu Key Laboratory of New Drug Research and Clinical Pharmacy. Dr Wang was the recipient of the 2019 Australia-China Helicobacter Research Fellowship (ACHRF) funded by the Australia-China Council, Department of Foreign Affairs and Trade, Australia.
Associate Professor Michael J Wise completed a double degree in Engineering and Arts and a PhD in electrical engineering at the University of New South Wales. He then worked for the University Technology, Sydney for two years before lecturing in Computer Science at the University of Sydney. There he created computer software for use in plagiarism detection, but then realised that his programs had an alternative application for sequence alignment, which prompted an interest in bioinformatics. Professor Wise was subsequently employed as a Senior Research Fellow at Pembroke College in Cambridge. In 2004 he moved to The University of Western Australia and discovered the world of microbes.


