Profiling bacterial communities in low biomass samples: pitfalls and considerations
Lisa F Stinson A C , Jeffrey A Keelan B and Matthew S Payne BA School of Molecular Sciences, The University of Western Australia, Perth, WA, Australia
B Division of Obstetrics and Gynaecology, Faculty of Health and Medical Sciences, The University of Western Australia, Perth, WA, Australia
C Tel: +61 6488 3200, Fax: +61 6488 7086, Email: lisa.stinson@uwa.edu.au
Microbiology Australia 40(4) 181-185 https://doi.org/10.1071/MA19053
Published: 14 November 2019
Bacterial 16S rRNA gene sequencing studies are popular across many fields of biology. This technique has allowed us to study bacterial communities like never before, leading to significant insights into microbial ecology and host– microbe interactions. However, 16S rRNA gene-based workflows are vulnerable to confounding and bias at every step. Many studies are plagued by entrenched methodological errors, producing data riddled with experimental artefacts. These issues are amplified in the study of low bacterial biomass samples, such as forensic and ancient samples, blood, meconium, ice and the built environment. It is, therefore, necessary to define the pitfalls of low biomass 16S rRNA gene-based work flows and to identify methods that may allow more accurate characterisation of bacterial communities in such samples.
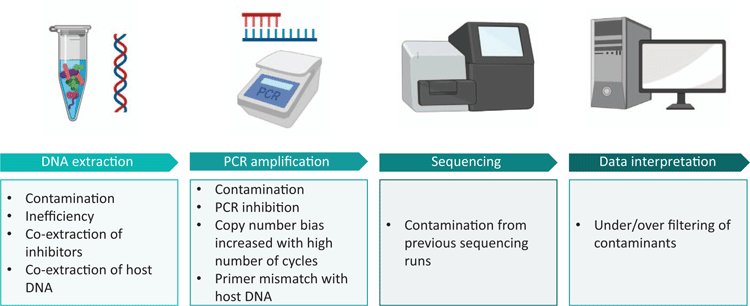
While the term ‘microbiome’ specifically relates to a microbial ecosystem present in a particular environment, and encompasses all microbes present (including bacteria, archaea, viruses, fungi, yeasts and protozoa), microbiome research remains heavily dominated by bacterial 16S rRNA gene sequencing studies. For this reason, the focus of this mini-review will be on the technical considerations associated with bacterial 16S rRNA gene profiling in low biomass environments (Figure 1).
Contamination
Low biomass samples are intrinsically vulnerable to contamination. The proportion of contaminating bacterial DNA increases with decreasing starting bacterial biomass1. Ultra-clean facilities, ultra-pure water and even certified DNA-free reagents harbour low levels of bacterial DNA2–7. It is, therefore, not possible to perform a microbiome experiment devoid of environmental contamination. External contamination may be introduced from reagents, laboratory surfaces and instruments or users. Sample-to-sample cross-contamination may be introduced from neighbouring wells or tubes when particles are aerosolised during pipetting, tube cap removal, plate seal removal or from centrifugation of open spin columns8. Further, contamination can occur on sequencing machines where residual barcodes or amplicons may be carried over from previous runs9,10. It is, therefore, crucial to take blank controls at sample collection, DNA extraction and PCR amplification. These controls should be sequenced alongside samples, even if no apparent amplification is present post-PCR, and taken into account during data interpretation.
Bacterial DNA introduced from DNA extraction kits (the ‘kitome’) and PCR master mixes (the ‘mixome’) have both been shown to significantly contribute to contamination11,12. The extent to which one or the other will dominate the contamination profile will vary between labs. We have recently shown that the mixome was the major source of contamination in our own workflow12. Importantly, we have been able to reduce the mixome by 99% using a commercially available dsDNase kit12. Enzymatic decontamination of PCR reagents should be incorporated into all 16S rRNA gene sequencing workflows, especially those of low biomass, to minimise reagent-based contamination.
Numerous studies have shown that different batches of the same reagents will introduce different contamination profiles3,11,13,14. Batch numbers should therefore be recorded for each step, and batch effects should be planned for when processing samples. For example, if blood samples from patients with a particular disease are compared to those without that disease, these should be randomised within a batch. If they are processed in separate batches resulting differences between their bacterial DNA profiles may be mere artefacts of the batch effect. Batch-specific contamination should be identifiable in negative controls, again reiterating the importance of prudent use of controls in low biomass work. While bioinformatic tools are available for detecting batch effects14, the simple use of appropriate negative controls and sample randomisation are sufficient.
Differentiating live from dead
DNA-based analysis of bacterial communities is broadly useful for characterising the taxa present; however, it is unable to differentiate live, metabolically active cells from dead cells or cell-free DNA. We have previously demonstrated the importance of excluding non-viable bacteria when analysing bacterial profiles in low biomass samples15. The presence of cell-free DNA and non-viable bacteria in low biomass samples can significantly confound the biological interpretation of sequence data. Exclusion of DNA from non-viable bacteria should, therefore, be integrated into low biomass 16S rRNA gene sequencing work flows. While there are numerous approaches for distinguishing live bacteria from dead16, the use of viability dyes is the most compatible with 16S rRNA gene sequencing work. Viability dyes, such as ethidium monoazide (EMA) and propidium monoazide (PMA), are cell membrane impermeable DNA intercalating agents17. These dyes are able to pass through the compromised cell membranes of dead bacteria, where they intercalate with the DNA inside, and upon photo-activation become covalently cross-linked to it. This strongly inhibits PCR amplification of affected DNA molecules, resulting in the amplification of DNA derived from intact cells only. Viability dyes have previously been used to exclude non-viable bacteria in the analysis of low biomass environments such as meconium, cleanrooms and even the international space station15,18–20. However, viability dyes are imperfect, as their effectiveness varies with different bacterial species and in different sample types17. Their use requires optimisation and validation for the sample type at hand.
Host DNA
In the analysis of clinical samples, host DNA is a double-edged sword. On one hand, 16S rRNA gene primers can mismatch with host DNA, creating a false positive result on a bacterial DNA presence/absence level and consuming valuable reagents in downstream sequencing applications, thus greatly limiting interpretation of resultant data21. Alternatively, host DNA may act as a PCR inhibitor, creating a false negative result22. A number of commercial kits (such as MolYsis) and bespoke methods (osmotic lysis, Saponin) have been trialled for depletion of host DNA with varying levels of success23,24. Host depletion generally consists of selective (mammalian) cell lysis followed by DNase treatment. However, such methods tend to rely on high microbial load and high initial sample volume and may therefore be unsuitable for low biomass samples23,25,26. Furthermore, pre-lysis steps may inadvertently lyse some bacterial cells (such as Ureaplasma spp.) and thereby distort the resulting bacterial DNA profiles. In the context of low biomass clinical samples, it is worth measuring host DNA levels using targets such as human β-globin to ensure that a negative result is not a consequence of sample insufficiency.
PCR inhibitors
The presence of PCR inhibitors in low biomass samples may create a false negative result. Common PCR inhibitors include urea, haemoglobin, and bile in clinical samples, and humic acids, polysaccharides and minerals in environmental samples27. A frequently used method for reducing the effects of PCR inhibitors is dilution of the sample. However, when the quantity of template DNA is low to begin with, dilution may reduce the template to undetectable levels. It is, therefore, important to select appropriate methods for DNA extraction and PCR amplification. In our own experience, we have found that increasing the standard ‘inhibitor removal’ steps in commercial extraction kits can reduce the co-extraction of PCR inhibitors28. Further, different DNA polymerases have varying abilities to perform in the presence of PCR inhibitors29. PCR reagents should therefore be selected based on the inhibitors expected to be present in the sample type being processed. In general, the effect of PCR inhibition in low biomass samples can be assessed by spiking a known concentration of pure bacterial DNA from a species that is unlikely to be found in the sample type (positive control) into the extracted sample DNA. This allows more accurate interpretation of ostensibly ‘negative’ results in low biomass settings.
Data interpretation
Data produced from low biomass samples should be interpreted very cautiously, particularly where interpretations of sterility are being made. One perk of low biomass work is the fact that samples often contain a very low diversity of reads, making manual examination of sequences unlaborious. Often, common sense identification of ‘blue whales in the Himalayas or African elephants in Antarctica’14 is useful in recognising potential contaminants. For instance, in our recent publication examining fetal bacterial communities, we identified two OTUs belonging to the thermophilic taxa Thermothrix azorensis and Thermus scotoductus, which were unlikely human microbiome candidates30. The inclusion of contaminating sequences in bacterial analysis of low biomass samples can distort the apparent composition of a sample and inflate diversity measures1. Karstens et al. compared four methods for filtering contaminating reads from low biomass data sets (removing all sequences present in a negative control, removing low abundance sequences, removing sequences that have an inverse correlation with DNA concentration31, and using SourceTracker to predict sequences arising from defined contaminant sources)1. These methods varied in their ability to delineate contaminants from true sequences. Notably, removing all sequences present in negative controls erroneously removed >20% of expected sequences. SourceTracker was able to remove contaminating sequences with a high level of accuracy when the experimental environment was well defined, but performed very poorly when the experimental environment was unknown. The most successful method (identifying sequences that have an inverse correlation with DNA concentration using the R package Decontam31) was still only able to identify 70–90% of contaminants. It is, therefore, vital to decontaminate reagents and to adopt working habits that minimise user- or environmentally introduced contamination. As the golden rule of microbiome research states: rubbish in, rubbish out.
Robust practices to avoid spurious conclusions
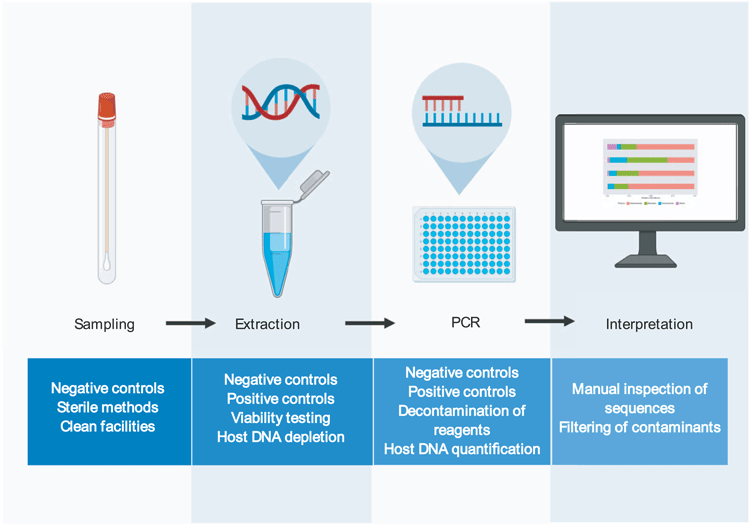
Here we will put forward a series of suggested methods that will minimise the pitfalls associated with low biomass 16S rRNA gene sequencing workflows (Figure 2).
Sample collection, handling, and facilities
Blank controls should be taken at the point of sample collection and taken through to sequencing to characterise any contamination introduced by the sampling tubes/swabs/environment. Samples and tubes should only ever be handled with gloves to avoid contamination from skin bacteria. It is important to note that specialised laboratories are optimal for low biomass work. These may include specialised clean room facilities, or even simple measures such as having separate labs for DNA extraction (raw samples only), template preparation (raw DNA only, no amplicons) and PCR (amplicons). Regardless of the facilities being used, personnel should always wear disposable gloves and lab coats and work within laminar flow cabinets. All work surfaces should be decontaminated before and after use with 10% bleach and UV irradiation. All plastic-ware used should be certified DNA-free, and pipette tips should be filtered. These basic steps are crucial for minimising lab-ware- and user-introduced contamination.
Sample preparation
Depending on the sample type, host DNA depletion may be necessary prior to DNA extraction. Viability dyes may also be used prior to DNA extraction to differentiate viable and non-viable bacteria.
DNA extraction
It is important to take negative extraction controls with each batch of extractions and to record where this control sits in relations to other samples. Extraction methods should be optimised for the sample type being used. Some sample types may require optimisation of lysis or inhibitor removal. For example, many commercially available kits are unable to extract DNA from meconium without further optimisation28. These samples may, therefore, be incorrectly deemed sterile if not properly extracted. Internal extraction controls can be spiked in to samples to quantify extraction efficiency13. However, it should be noted that the use of exogenous bacteria as an internal control cannot provide information on the efficiency of an extraction method for lysing/purifying bacteria that is endogenous to a sample. Importantly, spiking bacterial cells into a low biomass sample introduces the risk that the internal control will out-number the bacteria that are truly present, thereby sequestering the majority of the reads in a sequencing run.
PCR
Enzymatic decontamination of PCR reagents is a simple and effective method for reducing contamination12. Negative PCR controls should be used and sequenced to characterise any contamination introduced during this step. Again, the position of these controls on the plate in relation to other samples should be recorded to control for well-to-well contamination. Internal positive controls may be used at this stage to detect any PCR inhibition.
Data interpretation
Data should be interpreted with caution. Potential contaminants should be openly reported and post-hoc methods to remove contaminating reads should be utilised to increase the accuracy of downstream analysis. However, care must be taken in selecting methods for removing contaminating sequences, as overzealous filtering of potential contaminants can lead to spurious conclusions32.
Conclusions
16S rRNA gene sequencing technologies have allowed extensive characterisation of bacterial communities across numerous environments. These technologies are far more sensitive than previous culture-based methods of bacterial profiling, allowing the detection of very low titres of bacterial DNA from complex samples. However, findings from low biomass 16S rRNA gene sequencing studies can be undermined by reagent-based contamination and other methodological issues. There is, therefore, a need to adopt robust and conservative approaches to study low biomass bacterial communities.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work did not receive any specific funding.
References
[1] Karstens, L. et al. (2019) Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems 4, e00290-19.| Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments.Crossref | GoogleScholarGoogle Scholar | 31164452PubMed |
[2] Weyrich, L.S. et al. (2019) Laboratory contamination over time during low-biomass sample analysis. Mol. Ecol. Resour. 19, 982–996.
| Laboratory contamination over time during low-biomass sample analysis.Crossref | GoogleScholarGoogle Scholar | 30887686PubMed |
[3] Glassing, A. et al. (2016) Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 8, 24.
| Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples.Crossref | GoogleScholarGoogle Scholar | 27239228PubMed |
[4] Weiss, S. et al. (2014) Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 15, 564.
| Tracking down the sources of experimental contamination in microbiome studies.Crossref | GoogleScholarGoogle Scholar | 25608874PubMed |
[5] Grahn, N. et al. (2003) Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol. Lett. 219, 87–91.
| Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons.Crossref | GoogleScholarGoogle Scholar | 12594028PubMed |
[6] Kulakov, L.A. et al. (2002) Analysis of bacteria contaminating ultrapure water in industrial systems. Appl. Environ. Microbiol. 68, 1548–1555.
| Analysis of bacteria contaminating ultrapure water in industrial systems.Crossref | GoogleScholarGoogle Scholar | 11916667PubMed |
[7] McAlister, M.B. et al. (2002) Survival and nutritional requirements of three bacteria isolated from ultrapure water. J. Ind. Microbiol. Biotechnol. 29, 75–82.
| Survival and nutritional requirements of three bacteria isolated from ultrapure water.Crossref | GoogleScholarGoogle Scholar | 12161774PubMed |
[8] Minich, J.J. et al. (2019) Quantifying and understanding well-to-well contamination in microbiome research. mSystems 4, e00186-19.
| Quantifying and understanding well-to-well contamination in microbiome research.Crossref | GoogleScholarGoogle Scholar | 31239396PubMed |
[9] de Goffau, M.C. et al. (2019) Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334.
| Human placenta has no microbiome but can contain potential pathogens.Crossref | GoogleScholarGoogle Scholar | 31367035PubMed |
[10] Seitz, V. et al. (2015) A new method to prevent carry-over contaminations in two-step PCR NGS library preparations. Nucleic Acids Res. 43, e135.
| A new method to prevent carry-over contaminations in two-step PCR NGS library preparations.Crossref | GoogleScholarGoogle Scholar | 26152304PubMed |
[11] Salter, S.J. et al. (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87.
| Reagent and laboratory contamination can critically impact sequence-based microbiome analyses.Crossref | GoogleScholarGoogle Scholar | 25387460PubMed |
[12] Stinson, L.F. et al. (2019) Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass microbiome analyses. Lett. Appl. Microbiol. 68, 2–8.
| Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass microbiome analyses.Crossref | GoogleScholarGoogle Scholar | 30383890PubMed |
[13] Eisenhofer, R. et al. (2019) Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27, 105–117.
| Contamination in low microbial biomass microbiome studies: issues and recommendations.Crossref | GoogleScholarGoogle Scholar | 30497919PubMed |
[14] de Goffau, M.C. et al. (2018) Recognizing the reagent microbiome. Nat. Microbiol. 3, 851–853.
| Recognizing the reagent microbiome.Crossref | GoogleScholarGoogle Scholar | 30046175PubMed |
[15] Stinson, L.F. et al. (2019) Characterization of the bacterial microbiome in first-pass meconium using propidium monoazide (PMA) to exclude nonviable bacterial DNA. Lett. Appl. Microbiol. 68, 378–385.
| Characterization of the bacterial microbiome in first-pass meconium using propidium monoazide (PMA) to exclude nonviable bacterial DNA.Crossref | GoogleScholarGoogle Scholar | 30674082PubMed |
[16] Emerson, J.B. et al. (2017) Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5, 86.
| Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems.Crossref | GoogleScholarGoogle Scholar | 28810907PubMed |
[17] Fittipaldi, M. et al. (2012) Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91, 276–289.
| Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification.Crossref | GoogleScholarGoogle Scholar | 22940102PubMed |
[18] Vaishampayan, P. et al. (2013) New perspectives on viable microbial communities in low-biomass cleanroom environments. ISME J. 7, 312–324.
| New perspectives on viable microbial communities in low-biomass cleanroom environments.Crossref | GoogleScholarGoogle Scholar | 23051695PubMed |
[19] Weinmaier, T. et al. (2015) A viability-linked metagenomic analysis of cleanroom environments: eukarya, prokaryotes, and viruses. Microbiome 3, 62.
| A viability-linked metagenomic analysis of cleanroom environments: eukarya, prokaryotes, and viruses.Crossref | GoogleScholarGoogle Scholar | 26642878PubMed |
[20] Checinska Sielaff, A. et al. (2019) Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome 7, 50.
| Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces.Crossref | GoogleScholarGoogle Scholar | 30955503PubMed |
[21] Kommedal, Ø. et al. (2012) Dual priming oligonucleotides for broad-range amplification of the bacterial 16S rRNA gene directly from human clinical specimens. J. Clin. Microbiol. 50, 1289–1294.
| Dual priming oligonucleotides for broad-range amplification of the bacterial 16S rRNA gene directly from human clinical specimens.Crossref | GoogleScholarGoogle Scholar | 22278843PubMed |
[22] Radomski, N. et al. (2013) The critical role of DNA extraction for detection of mycobacteria in tissues. PLoS One 8, e78749.
| The critical role of DNA extraction for detection of mycobacteria in tissues.Crossref | GoogleScholarGoogle Scholar | 24194951PubMed |
[23] Bachmann, N.L. et al. (2018) Advances in clinical sample preparation for identification and characterization of bacterial pathogens using metagenomics. Front. Public Health 6, 363.
| Advances in clinical sample preparation for identification and characterization of bacterial pathogens using metagenomics.Crossref | GoogleScholarGoogle Scholar | 30619804PubMed |
[24] Marotz, C.A. et al. (2018) Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome 6, 42.
| Improving saliva shotgun metagenomics by chemical host DNA depletion.Crossref | GoogleScholarGoogle Scholar | 29482639PubMed |
[25] Doughty, E.L. et al. (2014) Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ 2, e585.
| Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer.Crossref | GoogleScholarGoogle Scholar | 25279265PubMed |
[26] Votintseva, A.A. et al. (2017) Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 55, 1285–1298.
| Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples.Crossref | GoogleScholarGoogle Scholar | 28275074PubMed |
[27] Schrader, C. et al. (2012) PCR inhibitors – occurrence, properties and removal. J. Appl. Microbiol. 113, 1014–1026.
| PCR inhibitors – occurrence, properties and removal.Crossref | GoogleScholarGoogle Scholar | 22747964PubMed |
[28] Stinson, L.F. et al. (2018) Comparison of meconium DNA extraction methods for use in microbiome studies. Front. Microbiol. 9, 270.
| Comparison of meconium DNA extraction methods for use in microbiome studies.Crossref | GoogleScholarGoogle Scholar | 29515550PubMed |
[29] Al-Soud, W.A. and Radstrom, P. (2001) Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39, 485–493.
| Purification and characterization of PCR-inhibitory components in blood cells.Crossref | GoogleScholarGoogle Scholar | 11158094PubMed |
[30] Stinson, L.F. et al. (2019) The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front. Microbiol. 10, 1124.
| The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth.Crossref | GoogleScholarGoogle Scholar | 31231319PubMed |
[31] Davis, N.M. et al. (2018) Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226.
| Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data.Crossref | GoogleScholarGoogle Scholar | 30558668PubMed |
[32] Payne, M.S. et al. (2019) Re: ‘Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community’ (2018) 6:87, https://doi.org/10.1186/s40168-018-0475-7. Microbiome 7, 20.
| Re: ‘Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community’ (2018) 6:87, https://doi.org/10.1186/s40168-018-0475-7.Crossref | https://doi.org/10.1186/s40168-018-0475-7.&journal=Microbiome&volume=7&pages=20-&publication_year=2019&author=M%2ES%2E%20Payne&hl=en&doi=10.1186/s40168-019-0642-5" target="_blank" rel="nofollow noopener noreferrer" class="reftools">GoogleScholarGoogle Scholar | 30755258PubMed |
Biographies
Lisa Stinson is a Research Fellow in the Hartmann Human Lactation Research Group at the University of Western Australia. She recently completed her PhD studying the human fetal microbiome. Her research now focuses on the human milk microbiome.
Jeffrey Keelan has 30 years’ experience in pregnancy research, focusing on placental inflammation, intrauterine infection and preterm birth. He is Head of Laboratories at the Division of Obstetrics and Gynaecology and Head of the School of Biomedical Sciences at the University of Western Australia.
Matthew Payne is a Senior Research Fellow within the School of Medicine at the University of Western Australia, and a highly experienced molecular microbiologist with expertise in perinatal microbiology.