Thiocyanate biodegradation: harnessing microbial metabolism for mine remediation
Mathew P Watts A and John W Moreau A BA School of Earth Sciences, The University of Melbourne, Parkville, Vic., Australia
B Email: jmoreau@unimelb.edu.au
Microbiology Australia 39(3) 157-161 https://doi.org/10.1071/MA18047
Published: 7 August 2018
Thiocyanate (SCN–) forms in the reaction between cyanide (CN–) and reduced sulfur species, e.g. in gold ore processing and coal-coking wastewater streams, where it is present at millimolar (mM) concentrations1. Thiocyanate is also present naturally at nM to µM concentrations in uncontaminated aquatic environments2. Although less toxic than its precursor CN–, SCN– can harm plants and animals at higher concentrations3, and thus needs to be removed from wastewater streams prior to disposal or reuse. Fortunately, SCN– can be biodegraded by microorganisms as a supply of reduced sulfur and nitrogen for energy sources, in addition to nutrients for growth4. Research into how we can best harness the ability of microbes to degrade SCN– may offer newer, more cost-effective and environmentally sustainable treatment solutions5. By studying biodegradation pathways of SCN– in laboratory and field treatment bioreactor systems, we can also gain fundamental insights into connections across the natural biogeochemical cycles of carbon, sulfur and nitrogen6.
Thiocyanate: a common wastewater contaminant
Thiocyanate (SCN–) is a common contaminant associated with a range of industries, and is typically found at its highest concentrations in the wastewater of gold and silver mines as a result of the use of CN– as a lixiviant during ore processing7. Thiocyanate is also commonly a component of coal coking wastewater alongside phenol and CN–8. These typically voluminous waste streams pose a serious environmental hazard due to their persistence and toxicity. Although a number of chemical SCN– degradation techniques exist, they are often inefficient and expensive9. Alternatively, many mines choose to store contaminated tailings indefinitely in dam structures, and re-use the SCN– contaminated water during ore processing. However, the presence of SCN– in this re-used water is known to impact gold extraction efficiency negatively10, as well as to impede the metabolism of biomining microorganisms11. Improved SCN– treatment systems, therefore, offer an opportunity improve the sustainability of mining processes globally.
Diversity of thiocyanate-degrading microorganisms
Thiocyanate offers a rich source of growth nutrients and energy to microorganisms, in the form of reduced sulfur and nitrogen, and a number of microbial species are known to be capable of SCN– degradation4. Importantly, these species do not belong to a distinct phylogenetic group, and the presence/absence of SCN–-degrading potential is often even strain specific. This complicates their identification using phylogenetic markers, such as the 16S rRNA gene. Much of what is currently known has, therefore, been achieved through culturing experiments. These studies have revealed diverse metabolic traits associated with SCN– degradation, where chemolithotrophs utilise the reduced sulfur as an energy source12–14, and heterotrophs utilise the nitrogen as a growth nutrient15–17. Despite SCN– biodegradation being widely regarded as an aerobic process, one bacterium (Thialkalivibrio thiocyanodenitrificans) was found to be able to couple this process to nitrate reduction14, opening up the possibility of anaerobic SCN– degradation. Experimentation on culturable strains has also allowed the elucidation of two distinct pathways of SCN– degradation. These pathways proceed via two intermediates, carbonyl sulfide (COS) or cyanate (CNO–), catalysed by one of two distinct types of SCN– hydrolases (SCNase)18,19, or by an SCN– dehydrogenase (TcDH) enzyme20,21, respectively. The resulting COS and CNO– intermediate chemical species are then available for degradation by enzymes associated with β-carbonic hydrase22 or cyanate anhydrase23. Both of these SCN– biodegradation pathways result in the release of reduced sulfur (S2– or S0), NH4+ and CO2.
The advent of high-throughput sequencing, and the rise in genome/metagenome and proteome/metaproteome sequences, has yielded vast databases of protein and DNA sequences. Significantly, protein sequences for the three known SCN–-degrading enzymes are available, which allow a deeper look into the distribution of SCN–-degrading enzymes. The three-subunit SCNase enzyme, originally isolated in Thiobacillus thioparus THI11518, is the most widely identified enzyme in protein databases, with 416 sequences identified as the γ-subunit in the NCBI nr database. These sequences primarily belong to the Actinobacteria, due to a bias towards full genome sequences of the medically significant Mycobacterium genus. They also contain sequences belonging to a number of thiobacilli, sulfur-oxidising γ-proteobacteria (including a number of Chromatiales) and α-proteobacteria (Methylobacterium spp. and Sphingomonas spp.). The alternative SCNase and the TcDH have far fewer closely related sequences in the NCBI non-redundant protein sequence database. This SCNase has representatives encoded in a number of thiobacilli, Afipia spp. and sulfur-oxidising γ-proteobacteria. The TcDH, originally purified from two Thioalkalivibrio spp.21, appears to be closely related to other Thioakalivibrio, and more distantly related to proteins of unknown function in Thioploca ingrica, Nitrospirae and Hydrogenobacter thermophilus. The comparatively few metaproteome and metagenome studies, targeting SCN–-contaminated systems, limit our understanding of the true scope of the distribution of these enzymes.
Thiocyanate biodegradation triggers complex microbial community interactions
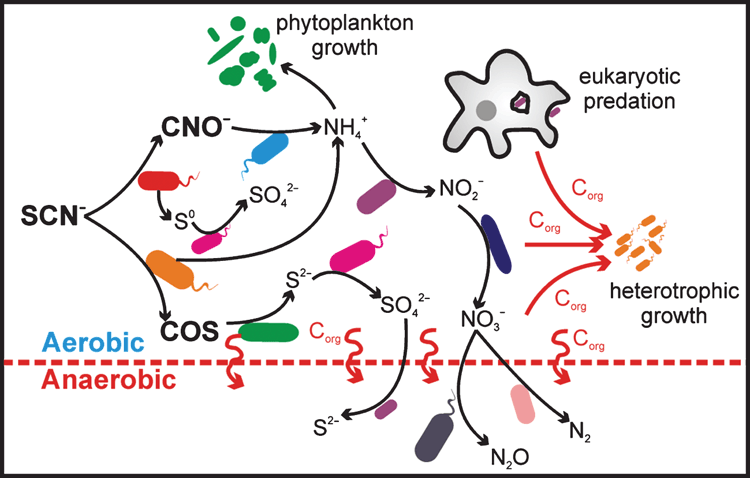
Although SCN– degradation is undertaken by a limited number of bacterial species (or strains), wider implications can result for the whole microbial community due to the roles its constituent elements can play as biological energy sources or growth nutrients. As a result, interesting and potentially useful symbiotic or dependent relationships can develop between SCN– degraders and non-SCN– degraders (Figure 1).
Thiocyanate-degrading bioreactors are often dominated by SCN–-degrading and sulfur-oxidising strains of the thiobacilli24,25. Although autotrophic, these bacteria have also been found to dominate in systems fed with a labile carbon source26,27. Their dominance is a testament to the strong selective pressure that SCN– exerts on a microbial community. The presence in these systems of a number of sulfur-oxidising taxa that do not genetically encode for known SCN–-degrading enzymes suggests a fraction of the reduced sulfur is shared within the community. The eventual liberation of NH4+ from SCN– degradation also opens up potential cross-feeding relationships. Several SCN–-treating systems were found to sustain populations of nitrifying bacteria, utilising NH4+ as an energy source and lacking the ability to degrade SCN–24,26. The subsequent nitrate can also supply an electron acceptor for denitrification in anoxic regions of these systems26. The supply of NH4+ as a nitrogen-containing nutrient likely provides the primary source of nitrogen for growth of microbial communities inhabiting these systems25. This nitrogen can also support phytoplankton growth under phototrophic conditions28. These symbiotic relationships have the potential to be harnessed for the effective removal of nitrogen from wastewater following SCN– degradation.
As SCN– degradation is able to support dominant populations of chemolithotrophs, it has the potential to exert a large influence on carbon cycling in SCN– treatment systems. Indeed, the SCN–-degrading thiobacilli and Thioalkalivibrio are known to utilise the Calvin-Benson-Bassham cycle to fix CO221,26. In the absence of a labile carbon source, the sulfur- and nitrogen-oxidising chemolithotrophs can provide the primary source of carbon in the system. Our own work, for example, found that a SCN– treatment system operated without labile carbon input, and was dominated by chemolithotrophs, sustaining SCN– degradation and a considerable diversity of heterotrophic microorganisms25. This study also implicated a role for bacterivorous Amoebazoa in preying on the microbial community and closing the microbial loop in the system.
Implications for thiocyanate-degrading biotechnology
Microbial communities capable of SCN– degradation offer an opportunity to treat large quantities of wastewater effectively. Such systems, adopting various designs, have already been deployed with success at field scale24,29,30. Much research is now going into understanding these systems at the molecular scale, and developing more efficient processes. Understanding the aforementioned cross-feeding relationships within SCN– treatment systems are vitally important to this process improvement. A good example is the revelation, achieved through a genome-resolved metagenomics approach, that even in organic carbon-fed systems, the community can be dominated by SCN–-degrading autotrophic bacteria26. Our own work has proven experimentally that SCN–-degrading bioreactors can be operated in the absence of organic carbon amendments25. This autotrophic SCN– degradation approach, combined with nitrification and denitrification, has been deployed at pilot scale to treat contaminated groundwater at Stawell Gold Mines in Victoria (Figure 2). This process is able to completely degrade the influent SCN– (300–400 mg L–1), resulting in sulfate and N-containing products. By harnessing various metabolic niches the initially released NH4+ is nitrified to nitrate and subsequently removed by denitrification. In addition to this engineered approach, we reported for the first time the ability to promote in situ SCN– biodegradation in contaminated gold mine tailings water held in large open air storage facilities9. This process was promoted through phosphate nutrient addition alone, and offers a passive approach for the treatment of large quantities of contaminated material. Our future work aims to better resolve the active metabolic interactions within the microbial communities in these in situ and ex situ approaches. This will help us to promote beneficial symbiotic relationships within the system.
Collectively, the advances in our understanding of the metabolisms underpinning SCN– biodegradation allow for better designs and approaches to harnessing this microbial potential. Biodegradation of SCN–, therefore, is offering a route to improving the water efficiency of industrial processes such as gold mining on a global scale.
In conclusion, much has been learned at the molecular scale about biodegradation of SCN– since this metabolic trait was first reported in T. thioparus12, including the enzymes responsible and the community wide biogeochemical impacts. The advent of ‘multi-omics’ approaches is allowing us to probe these processes in situ in SCN–-biodegrading treatment systems. The few studies utilising these techniques have spurred significant process improvements, in addition to revealing fundamental insights into SCN– biodegradation and the subsequent metabolic cycling of its breakdown products. These insights, although gained from engineered systems, can help inform the global cycling of SCN–, CNO– and COS and better constrain the role of microbial communities in the wider carbon, nitrogen and sulfur cycles. Given the limited number of systems investigated to this depth, more work is needed to appreciate fully the diversity and complexity of microbial communities degrading SCN–.
Acknowledgements
This work was funded an Australian Research Council Linkage Program grant to J.W.M. (LP160100866) and co-funded by Stawell Gold Mines.
References
[1] Dash, R.R. et al. (2009) Cyanide in industrial wastewaters and its removal: a review on biotreatment. J. Hazard. Mater. 163, 1–11.| Cyanide in industrial wastewaters and its removal: a review on biotreatment.Crossref | GoogleScholarGoogle Scholar |
[2] Kurashova, I. et al. (2018) Kinetics of decomposition of thiocyanate in natural aquatic systems. Environ. Sci. Technol. 52, 1234–1243.
[3] Bhunia, F. et al. (2000) Toxicity of thiocyanate to fish, plankton, worm, and aquatic ecosystem. Bull. Environ. Contam. Toxicol. 64, 197–204.
| Toxicity of thiocyanate to fish, plankton, worm, and aquatic ecosystem.Crossref | GoogleScholarGoogle Scholar |
[4] Gould, W.D. et al. (2012) A critical review on destruction of thiocyanate in mining effluents. Miner. Eng. 34, 38–47.
| A critical review on destruction of thiocyanate in mining effluents.Crossref | GoogleScholarGoogle Scholar |
[5] Akcil, A. (2003) Destruction of cyanide in gold mill effluents: biological versus chemical treatments. Biotechnol. Adv. 21, 501–511.
| Destruction of cyanide in gold mill effluents: biological versus chemical treatments.Crossref | GoogleScholarGoogle Scholar |
[6] Watts, M.P. and Moreau, J.W. (2016) New insights into the genetic and metabolic diversity of thiocyanate-degrading microbial consortia. Appl. Microbiol. Biotechnol. 100, 1101–1108.
| New insights into the genetic and metabolic diversity of thiocyanate-degrading microbial consortia.Crossref | GoogleScholarGoogle Scholar |
[7] Mudder, T.I. et al. (2001) Chemistry and treatment of cyanidation wastes. Mining Journal Books, London, UK.
[8] Staib, C. and Lant, P. (2007) Thiocyanate degradation during activated sludge treatment of coke-ovens wastewater. Biochem. Eng. J. 34, 122–130.
| Thiocyanate degradation during activated sludge treatment of coke-ovens wastewater.Crossref | GoogleScholarGoogle Scholar |
[9] Watts, M.P. et al. (2017) In situ stimulation of thiocyanate biodegradation through phosphate amendment in gold mine tailings water. Environ. Sci. Technol. 51, 13353–13362.
| In situ stimulation of thiocyanate biodegradation through phosphate amendment in gold mine tailings water.Crossref | GoogleScholarGoogle Scholar |
[10] Guo, B. et al. (2014) Cyanide chemistry and its effect on mineral flotation. Miner. Eng. 66–68, 25–32.
| Cyanide chemistry and its effect on mineral flotation.Crossref | GoogleScholarGoogle Scholar |
[11] van Hille, R.P. et al. (2015) Effect of thiocyanate on BIOX® organisms: inhibition and adaptation. Miner. Eng. 75, 110–15.
[12] Happold, F.C. et al. (1958) Utilization of thiocyanate by Thiobacillus thioparus and T. thiocyanoxidans. Nature 182, 266–267.
| Utilization of thiocyanate by Thiobacillus thioparus and T. thiocyanoxidans.Crossref | GoogleScholarGoogle Scholar |
[13] Sorokin, D.Y. et al. (2014) Isolation and characterization of an obligately chemolithoautotrophic Halothiobacillus strain capable of growth on thiocyanate as an energy source. FEMS Microbiol. Lett. 354, 69–74.
| Isolation and characterization of an obligately chemolithoautotrophic Halothiobacillus strain capable of growth on thiocyanate as an energy source.Crossref | GoogleScholarGoogle Scholar |
[14] Sorokin, D.Y. et al. (2002) Thioalkalivibrio thiocyanoxidans sp. nov. and Thioalkalivibrio paradoxus sp. nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing bacteria capable of growth on thiocyanate, from soda lakes. Int. J. Syst. Evol. Microbiol. 52, 657–664.
| Thioalkalivibrio thiocyanoxidans sp. nov. and Thioalkalivibrio paradoxus sp. nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing bacteria capable of growth on thiocyanate, from soda lakes.Crossref | GoogleScholarGoogle Scholar |
[15] Stratford, J. et al. (1994) The utilization of thiocyanate as a nitrogen source by a heterotrophic bacterium: the degradative pathway involves formation of ammonia and tetrathionate. Microbiology 140, 2657–2662.
| The utilization of thiocyanate as a nitrogen source by a heterotrophic bacterium: the degradative pathway involves formation of ammonia and tetrathionate.Crossref | GoogleScholarGoogle Scholar |
[16] Betts, P.M. et al. (1979) Thiocyanate utilization by an Arthrobacter. Can. J. Microbiol. 25, 1277–1282.
| Thiocyanate utilization by an Arthrobacter.Crossref | GoogleScholarGoogle Scholar |
[17] Wood, A.P. et al. (1998) A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch. Microbiol. 169, 148–158.
| A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources.Crossref | GoogleScholarGoogle Scholar |
[18] Katayama, Y. et al. (1992) A thiocyanate hydrolase of Thiobacillus thioparus. A novel enzyme catalyzing the formation of carbonyl sulfide from thiocyanate. J. Biol. Chem. 267, 9170–9175.
[19] Hussain, A. et al. (2013) Cloning and expression of a gene encoding a novel thermostable thiocyanate-degrading enzyme from a mesophilic alphaproteobacteria strain THI201. Microbiology 159, 2294–2302.
| Cloning and expression of a gene encoding a novel thermostable thiocyanate-degrading enzyme from a mesophilic alphaproteobacteria strain THI201.Crossref | GoogleScholarGoogle Scholar |
[20] Berben, T. et al. (2017) Analysis of the genes involved in thiocyanate oxidation during growth in continuous culture of the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio thiocyanoxidans ARh 2T using transcriptomics. MSystems 2, e00102-17.
| Analysis of the genes involved in thiocyanate oxidation during growth in continuous culture of the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio thiocyanoxidans ARh 2T using transcriptomics.Crossref | GoogleScholarGoogle Scholar |
[21] Berben, T. et al. (2017) Comparative genome analysis of three thiocyanate oxidizing thioalkalivibrio species isolated from soda lakes. Front. Microbiol. 8, 254.
[22] Ogawa, T. et al. (2016) Degradation of carbonyl sulfide by Actinomycetes and detection of clade D of β-class carbonic anhydrase. FEMS Microbiol. Lett. 363, fnw223.
| Degradation of carbonyl sulfide by Actinomycetes and detection of clade D of β-class carbonic anhydrase.Crossref | GoogleScholarGoogle Scholar |
[23] Anderson, P.M. et al. (1990) The cyanase operon and cyanate metabolism. FEMS Microbiol. Rev. 87, 247–252.
| The cyanase operon and cyanate metabolism.Crossref | GoogleScholarGoogle Scholar |
[24] Villemur, R. et al. (2015) Development of four-stage moving bed biofilm reactor train with a pre-denitrification configuration for the removal of thiocyanate and cyanate. Bioresour. Technol. 181, 254–262.
| Development of four-stage moving bed biofilm reactor train with a pre-denitrification configuration for the removal of thiocyanate and cyanate.Crossref | GoogleScholarGoogle Scholar |
[25] Watts, M.P. et al. (2017) Characterization of an autotrophic bioreactor microbial consortium degrading thiocyanate. Appl. Microbiol. Biotechnol. 101, 5889–5901.
[26] Kantor, R.S. et al. (2015) Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome‐resolved metagenomics. Environ. Microbiol. , .
| Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome‐resolved metagenomics.Crossref | GoogleScholarGoogle Scholar |
[27] Kantor, R.S. et al. (2017) Genome-resolved meta-omics ties microbial dynamics to process performance in biotechnology for thiocyanate degradation. Environ. Sci. Technol. 51, 2944–2953.
| Genome-resolved meta-omics ties microbial dynamics to process performance in biotechnology for thiocyanate degradation.Crossref | GoogleScholarGoogle Scholar |
[28] Ryu, B.-G. et al. (2015) A comprehensive study on algal–bacterial communities shift during thiocyanate degradation in a microalga-mediated process. Bioresour. Technol. 191, 496–504.
| A comprehensive study on algal–bacterial communities shift during thiocyanate degradation in a microalga-mediated process.Crossref | GoogleScholarGoogle Scholar |
[29] Whitlock, J.L. (1990) Biological detoxification of precious metal processing wastewaters. Geomicrobiol. J. 8, 241–249.
| Biological detoxification of precious metal processing wastewaters.Crossref | GoogleScholarGoogle Scholar |
[30] van Zyl, A.W. et al. (2011) Biodegradation of thiocyanate by a mixed microbial population. Mine Water–Managing the Challenges (IMWA 2011), Aachen, Germany.
Biographies
John W Moreau is a Senior Lecturer and Lead Investigator in the Geomicrobiology Lab at the School of Earth Sciences at The University of Melbourne. His main research interests include understanding the response of environmental microorganisms and microbial communities to metals and mining-related contaminants, and how to harness microbial activity to bioremediation strategies. He is also interested in studying the physiological and metabolic limitations of microbes thriving under extreme environmental conditions.
Mathew P Watts is a Postdoctoral Research Fellow in geomicrobiology at the School of Earth Sciences, The University of Melbourne. His primary research project focuses on understanding and developing a biological approach to thiocyanate remediation at a gold mine in Victoria. His general interests in the field extend to how microbial metabolic activities impact the form and fate of metals and organics in the environment.