The molecular epidemiology of Murray Valley encephalitis virus in Australasia
David T WilliamsCSIRO Australian Animal Health Laboratory
5 Portarlington Road
Geelong, Vic. 3220, Australia
Tel: +61 3 5227 5364
Email: D.Williams@csiro.au
Microbiology Australia 39(2) 106-108 https://doi.org/10.1071/MA18031
Published: 13 April 2018
Of the viruses transmitted by mosquitoes in the Australasian region, Murray Valley encephalitis (MVE) virus is the major cause of brain disease in humans. There is no vaccine to prevent MVE, nor are there effective antiviral drugs available to treat infections. Therefore, surveillance of MVE is essential to control efforts. A key element to this is understanding the virus at a genetic level, which allows the tracking and identification of known or novel genetic types and can tell us about their circulation patterns.
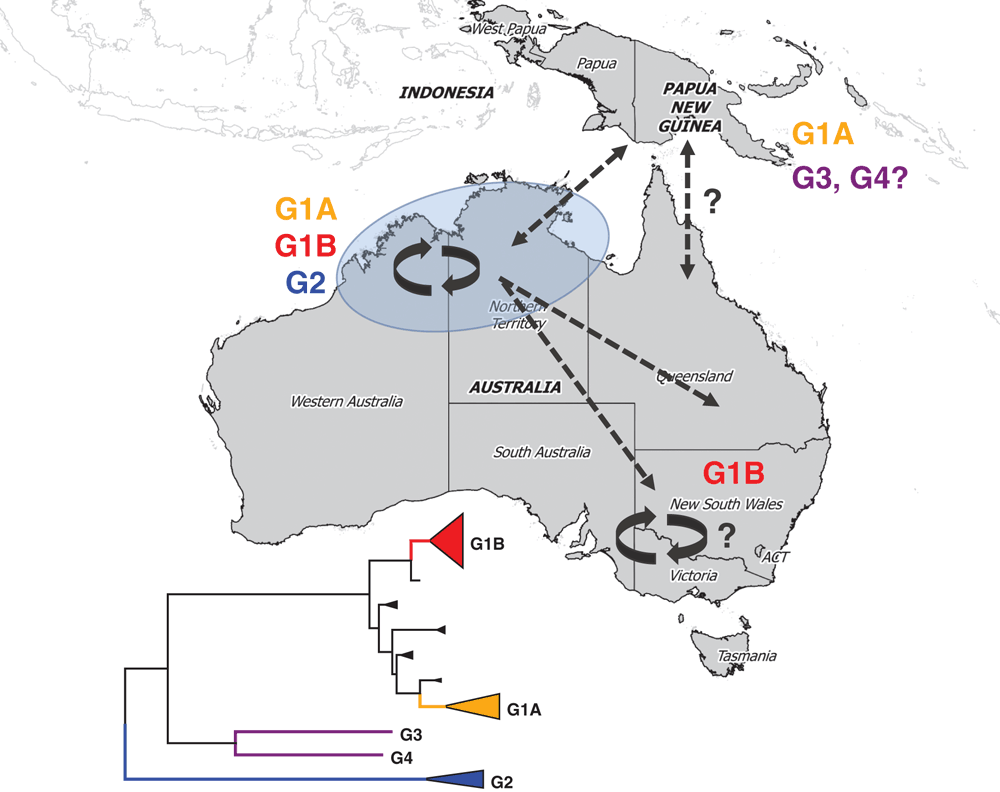
In the last century, the major epidemics of MVE occurred in southeastern Australia, centred on the Murray Valley region1. However, since 1978 the majority of cases have occurred in northern Western Australia and the Top End of the Northern Territory, where the virus is now endemic. Nowadays, MVE only occasionally re-emerges in south-eastern Australia following periods of heavy and prolonged rainfall. The last such event occurred in 2011, resulting in 17 cases of MVE nationwide and 3 deaths, including 2 cases (1 death) in South Australia and 1 case in New South Wales2. Approximately 100 equine cases were also reported in South Australia and the Eastern states in 20113. To the north of Australia, the virus is found in Papua New Guinea and Irian Jaya, where cases have been reported and virus isolated from mosquitoes1,4–6. MVE virus cycles between Culex species mosquitoes and water birds, such as herons and egrets1. It is not well understood how it reappears in places where occasional cases or activity is found, but it may be re-introduced by infected migratory water birds and/or wind-blown mosquitoes.
Four distinct genetic lineages or genotypes (G1-G4) of MVE virus have been identified (Figure 1), based on RNase oligonucleotide mapping5,7 or sequencing regions of the virus genome, such as the 5’ untranslated region or the structural genes pre-membrane and envelope8–10. Studies that analysed genes encoding the structural surface proteins of the virus have been the most comprehensive to date. As for other viruses, the surface proteins of MVE virus offer an attractive choice for phylogenetic analyses, since they are the primary target of the host immune response and subject to relatively strong selection pressures. This means that the genes encoding these proteins have high levels of sequence variation that allows sensitive levels of phylogenetic discrimination.
G1 is the major type of MVE virus on mainland Australia and the most recent to evolve from ancestral strains9,10. The last viruses identified from Papua New Guinea, isolated from mosquitoes caught in 1998, also belong to this type. Molecular clock analysis revealed that G1 evolved between 1930 and 194910. Within G1, two distinct sub-lineages (1A and 1B) have co-evolved and exhibit different patterns of transmission in Australia. G1A viruses were estimated to emerge in the 1970s and 80s from a common ancestor of early virus isolates from the 1950–51 and 1974 epidemics in southeast Australia. Paradoxically, Australian G1A viruses have only been found in the northwest of the country. The emergence of G1B in the 1980s overlapped that of G1A but, in contrast to the latter, G1B viruses have been found across mainland Australia (Figure 1). Both types have been associated with disease. We don’t yet know why they have different circulation patterns, but further studies may reveal subtle differences in the genome of G1B viruses that enables them to infect mosquitoes and waterbirds more efficiently, allowing them to outcompete their G1A counterparts.
|
Interestingly, the progenitor of G1B viruses emerged between 1968 and 197310, coinciding with construction of Stage 2 of the Ord River Irrigation scheme in the northeast Kimberley region of Western Australia. This involved damning the Ord River to create Lake Argyle, a vast freshwater reservoir, and resulted in profound changes to the local ecosystem. It has been hypothesised that this led to MVE virus becoming established in this region11. G1B subsequently became the dominant and most widespread lineage of MVE virus in Australia. It has been proposed that the changes to the ecosystem in the northeast Kimberley facilitated the emergence of this lineage, via increased populations of vector mosquitoes and waterbirds, and the creation of a permanent ecosystem suitable for maintaining mosquito-borne viruses10.
A second minor genotype (G2) is also found in Australia and consists of only a handful of mosquito isolates (~5% of all viruses propagated from trap-caught mosquitoes)9,12 and a single human isolate (J. Druce, personal communication), all from northwestern Australia. G2 is the oldest lineage of MVE virus, emerging directly from the ancestral virus of this species around 200 years ago10. Up until recently, it was thought that G2 viruses were attenuated, based on experiments using a mouse model of MVE10,13 and the absence of human cases linked to G2 viruses. However, the first isolation of a G2 virus from a case of MVE in the Northern Territory in 2015 indicates that G2 viruses have the same potential to cause severe disease as G1 viruses. It is not understood why G2 viruses are a minority type restricted to northwestern Australia. G2 strains may occupy a rarely-sampled ecological niche, or their genetic differences (to G1 viruses) may underlie altered biological properties that affect their replication and virulence, as for G1B viruses, thereby restricting their transmission9,10,12. In support of the latter, several unique amino acids are encoded in the major envelope surface glycoprotein that have potential biological significance.
The remaining two genotypes of MVE virus are ‘one hit wonders’ – single isolates from Papua New Guinea derived from a human case in 1956 (G3)4 and mosquitoes collected in 1966 (G4)5. No other strains of these genotypes have been found; however, surveillance activities in PNG and Irian Jaya have been limited and this may reflect under-sampling.
The co-circulation of all contemporary genotypes of MVE virus in northwestern Australia provides further evidence that this region is the enzootic focus for this virus. Continued surveillance throughout mainland Australia through existing mosquito surveillance programs, passive surveillance of cases and enhanced surveillance strategies will remain important to gather environmental and clinical samples to test for the presence of MVE viruses and gain a greater understanding of the molecular epidemiology of this potentially devastating virus. Although surveillance activities in PNG present challenges, the contribution to our knowledge of the epidemiology of MVE and other mosquito-borne diseases may be invaluable. Another gap for MVE virus molecular epidemiology is the lack of full length genomes. Currently, the complete genomes or ORFs of only seven different strains have been published14–16. Additional full length genomes will enable more comprehensive genetic studies that may uncover new insights into factors that determine transmission and pathogenicity. As next generation sequencing technologies become mainstream, this should be achievable in the near future.
References
[1] Marshall, I.D. (1988) Murray Valley and Kunjin encephalitis. In The arboviruses: epidemiology and ecology (Monath, T.P. ed), pp. 151–189, CRC Press, Boca Raton.[2] Selvey, L.A. et al. (2014) The changing epidemiology of Murray Valley encephalitis in Australia: the 2011 outbreak and a review of the literature. PLoS Negl. Trop. Dis. 8, e2656.
| The changing epidemiology of Murray Valley encephalitis in Australia: the 2011 outbreak and a review of the literature.Crossref | GoogleScholarGoogle Scholar |
[3] Roche, S.E. et al. (2013) Descriptive overview of the 2011 epidemic of arboviral disease in horses in Australia. Aust. Vet. J. 91, 5–13.
| Descriptive overview of the 2011 epidemic of arboviral disease in horses in Australia.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3szjtV2iug%3D%3D&md5=e1774fa0784be87e42569282ddef8e1eCAS |
[4] French, E.L. et al. (1957) Murray Valley encephalitis in New Guinea. I. Isolation of Murray Valley encephalitis virus from the brain of a fatal case of encephalitis occurring in a Papuan native. Am. J. Trop. Med. Hyg. 6, 827–834.
| Murray Valley encephalitis in New Guinea. I. Isolation of Murray Valley encephalitis virus from the brain of a fatal case of encephalitis occurring in a Papuan native.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaG1c%2Fht1ykuw%3D%3D&md5=4996484955ebf07e89ded1bb3d5bde16CAS |
[5] Lobigs, M. et al. (1986) Genetic differentiation of Murray Valley encephalitis virus in Australia and Papua New Guinea. Aust. J. Exp. Biol. Med. Sci. 64, 571–585.
| Genetic differentiation of Murray Valley encephalitis virus in Australia and Papua New Guinea.Crossref | GoogleScholarGoogle Scholar |
[6] Johansen, C.A. et al. (2000) Isolation of Japanese encephalitis virus from mosquitoes (Diptera: Culicidae) collected in the Western Province of Papua New Guinea, 1997–1998. Am. J. Trop. Med. Hyg. 62, 631–638.
| Isolation of Japanese encephalitis virus from mosquitoes (Diptera: Culicidae) collected in the Western Province of Papua New Guinea, 1997–1998.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M7pvFeitA%3D%3D&md5=5e821d1d0241b1f2e2b52484f4bfbf3dCAS |
[7] Coelen, R.J. and Mackenzie, J.S. (1988) Genetic variation of Murray Valley encephalitis virus. J. Gen. Virol. 69, 1903–1912.
| Genetic variation of Murray Valley encephalitis virus.Crossref | GoogleScholarGoogle Scholar |
[8] Coelen, R.J. and Mackenzie, J.S. (1990) The 5ʹ-terminal non-coding region of Murray Valley encephalitis virus RNA is highly conserved. J. Gen. Virol. 71, 241–245.
| The 5ʹ-terminal non-coding region of Murray Valley encephalitis virus RNA is highly conserved.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3cXhtlegtbY%3D&md5=c59010b058bf376a5f9b6f1fb3d1895cCAS |
[9] Johansen, C.A. et al. (2007) Genetic and phenotypic differences between isolates of Murray Valley encephalitis virus in Western Australia, 1972-2003. Virus Genes 35, 147–154.
| Genetic and phenotypic differences between isolates of Murray Valley encephalitis virus in Western Australia, 1972-2003.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXnvFeisbk%3D&md5=c8888b74f8d29585fba1e54b9f3d2ba8CAS |
[10] Williams, D.T. et al. (2015) The molecular epidemiology and evolution of Murray Valley encephalitis virus: recent emergence of distinct sub-lineages of the dominant genotype 1. PLoS Negl. Trop. Dis. 9, e0004240.
| The molecular epidemiology and evolution of Murray Valley encephalitis virus: recent emergence of distinct sub-lineages of the dominant genotype 1.Crossref | GoogleScholarGoogle Scholar |
[11] Mackenzie, J.S. and Broom, A.K. (1999) Ord River irrigation area: the effect of dam construction and irrigation on the incidence of Murray Valley encephalitis virus. In Water resources: Health, Environment and Development (Kay, B.H. ed), pp. 108–122, E & FN Spon.
[12] Niazi, A. (2014) The role of genetic diversity in the replication, pathogenecity and virulence of Murray Valley encephalitis virus, Curtin University, Perth, Western Australia.
[13] Coelen, R. (1988) Phenotypic and genotypic variation of Murray Valley encephalitis virus, Department of Microbiology, University of Western Australia, Perth, Western Australia.
[14] Hurrelbrink, R.J. et al. (1999) Characterization of infectious Murray Valley encephalitis virus derived from a stably cloned genome-length cDNA. J. Gen. Virol. 80, 3115–3125.
| Characterization of infectious Murray Valley encephalitis virus derived from a stably cloned genome-length cDNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXnvFyhu7g%3D&md5=7387038e0973e8d20bfb076fc7fb109fCAS |
[15] Mann, R.A. et al. (2013) Molecular characterization and phylogenetic analysis of Murray Valley encephalitis virus and West Nile virus (Kunjin subtype) from an arbovirus disease outbreak in horses in Victoria, Australia, in 2011. J. Vet. Diagn. Invest. 25, 35–44.
| Molecular characterization and phylogenetic analysis of Murray Valley encephalitis virus and West Nile virus (Kunjin subtype) from an arbovirus disease outbreak in horses in Victoria, Australia, in 2011.Crossref | GoogleScholarGoogle Scholar |
[16] Williams, D.T. et al. (2014) Complete genome sequences of the prototype isolates of genotypes 2, 3, and 4 of Murray Valley encephalitis virus. Genome Announc. 2, .
| Complete genome sequences of the prototype isolates of genotypes 2, 3, and 4 of Murray Valley encephalitis virus.Crossref | GoogleScholarGoogle Scholar |
[17] Kumar, S. et al. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874.
| MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhsF2ltrzN&md5=650eb206814988732cd47cfc1615513dCAS |
Biography
Dr David Williams is a Group Leader at the CSIRO Australian Animal Health Laboratory, Geelong, Victoria. His research and scientific interests are in the detection, diagnosis, and epidemiology of emerging and re-emerging viruses that affect humans and animals in Australia and overseas, including mosquito-borne viruses.


