Bat and virus ecology in a dynamic world
David A Wilkinson A and David TS Hayman A BA Molecular Epidemiology and Public Health Laboratory (mEpiLab), Hopkirk Research Institute, Massey University, Private Bag 11-222, Palmerston North, New Zealand
B Email: D.T.S.Hayman@massey.ac.nz
Microbiology Australia 38(1) 33-35 https://doi.org/10.1071/MA17011
Published: 9 February 2017
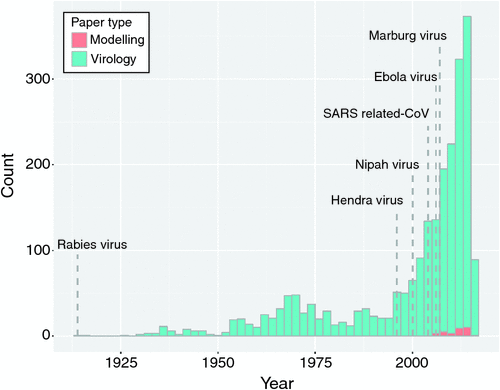
The emergence of infectious diseases caused by bat-associated viruses has had a devastating and wide-reaching effect on human populations. These viruses include lyssaviruses such as rabies virus, the filoviruses, Ebola (EBOV) and Marburg virus, Severe Acute Respiratory Syndrome (SARS) coronavirus, and the paramyxoviruses, Hendra virus (HeV) and Nipah virus (NiV)1. As a result bats have been the focus of substantial research (Fig. 1) and certain cellular and physiological traits of bats are hypothesised to lead to ‘special’ bat-virus associations2,3 (but see Han et al.4). The anthropogenic changes in the world we live will influence human health5, including through their impact on bat ecology and the viruses within bat populations. Australian people and livestock have been infected by novel bat viruses, such as HeV, Menangle viruses (MenV) and Australian bat lyssavirus (ABLV), and are at the forefront of both epidemiological and virological research efforts into cross-species transmission events (spillover): here we put some of those efforts and the potential impacts of anthropogenic changes on bat-virus ecology under the microscope.
|
The complex nature of bat virus spillover and disease emergence, encompassing environmental, ecological and biological factors, is perhaps best addressed through statistical and mathematical modelling frameworks that can integrate disparate data sources to make inferences regarding infection dynamics and risk6–8. Detailed epidemiological surveillance has helped to establish predictive models for disease transmission9. Recent work using HeV as a model has suggested a delineation of five major contributing factors to viral spillover: (1) reservoir host distribution and density; (2) pathogen shedding from reservoir hosts; (3) environmental stability of the pathogen; (4) recipient host exposure; and (5) recipient host susceptibility8. To successfully model these processes, we must understand reservoir community structures, the spatial and temporal nature of reservoir infection, reservoir host ranges and dispersal, as well as other factors that contribute to the ‘force of infection’ received by domestic animal or human populations10. Rich field-data is essential to fully understand and thus mitigate the risk from any bat associated zoonotic disease. Furthermore, as we progress through the Anthropocene era, bat populations meet new, direct and indirect challenges as a result of human population growth. These include climate change, habitat loss, increasing competition for resources and physical danger from man-made structures such as roads and wind-turbines11. Anthropogenic changes will alter bat population dynamics, infection dynamics within bat populations, and the contact rates between people, their animals, and bats. Each will affect the five major contributing factors to viral spillover8.
One of the most obvious anthropogenic changes is land-use change. With most land-use changes come non-native species, and it can be contact with non-native species that leads to spillover. Two obvious examples in Australia are HeV, which replicates and amplifies within horses12, and MenV, which infected pigs prior to people13. Both epidemiological and clinical reports highlight potential issues with emerging disease events. The first reported outbreak of HeV in Hendra, Brisbane, led clinicians to include poisoning, bacterial, viral, and other exotic diseases as potential causes before HeV was isolated12. In the case of MenV, the virus was isolated from stillborn piglets with deformities at a piggery in New South Wales in 1997. Stillbirth, birth defects, and mummified foetuses are not uncommon in production systems, so determining when novel infections are the cause requires appropriate microbiological studies. Such spillover events are rare, but whether direct human infection is likely, for either HeV or MenV, without intermediate hosts is unknown and prevention of HeV emergence focuses on stopping HeV infection in horses14. How infections then persist in novel host populations to facilitate human spillover, such as for NiV in pigs15, and how they may then perpetuate in human populations, such as NiV16 or EBOV17 in people, is then determined by a number of other virological and ecological processes.
Land-use change also typically leads to native habitat loss and fragmentation. Both processes may change bat behaviour and distribution. In doing so, these changes will inevitably affect the infection dynamics of the viruses within those populations. Plowright et al. modelled the transmission dynamics of HeV in Australian Pteropus bats (fruit bats or flying foxes)18. They developed an ecological model of the HeV dynamics within the populations and then used field and laboratory data to provide values for the parameters. The models included urban habituation with decreased migration of bat sub-populations, an observed change in Australian fruit bat ecology. Models predicted that decreased bat movement could lead to a decline in population immunity through reduced transmission among sub-populations, giving rise to more intense outbreaks after HeV reintroduction. Furthermore, analyses of spatially varying Eucalypt vegetation indices and weather events suggest ≥50% of landscape-scale bat behaviour is driven by Eucalypt resources19. These results suggest land-use changes interact with bat behaviour to determine HeV risk. Habitat fragmentation has been linked to other bat-related diseases, including NiV encephalitis20 and EBOV disease21. In general, land-use changes largely affect reservoir host distribution and density, potentially pathogen shedding from reservoir hosts and contact with novel recipient species. Once better characterised, predictive models of land-use change may increase our understanding of risk, and allow the development of strategies to mitigate the risk of spillover.
The impact of land-use change, including introduced species, is relatively well characterised for mammalian species, though not for the viruses within them. However, other anthropogenic changes that impact mammalian hosts remain poorly characterised, for example; anthropogenic climate change may impact other aspects of viral-host dynamics and therefore spillover risk. Plowright et al. performed a longitudinal study of HeV in little red flying foxes (Pteropus scapulatus)22. Serological data showed that pregnant and lactating females had significantly higher risk of infection and implied that HeV is transmitted horizontally and immunity (inferred through serology) wanes rapidly. The highest seroprevalence, however, was observed when bats were nutritionally stressed. Stress may reduce the capacity of bats to respond to viral infections through innate immune pathways2,23, leading to greater viral replication and adaptive immune responses. Martin et al. suggest that the bioclimatic niche of two species (P. alecto and P. conspicillatus) determines the spatial pattern of spillover of HeV24, whereas the environmental survival of HeV was limited in ‘normal’ environmental conditions25. These findings suggest anthropogenic changes that increase stress, such as habitat loss and climate change, may increase HeV risk in the future.
How generalisable these Australian study findings are is yet to be determined, but from an international perspective Australian bat-viral systems have been useful models to help understand bat-viral ecology and its interaction with anthropogenic change. The findings can potentially help reduce the threat of viral emergence in our dynamic world, and thus save lives, provided that lessons are learned from them and that sufficient and appropriate action is taken to mitigate the drivers of viral spillover.
References
[1] Hayman, D.T.S. (2016) Bats as viral reservoirs. Ann. Rev. Virol. 3, 1–609.| Bats as viral reservoirs.Crossref | GoogleScholarGoogle Scholar |
[2] Brook, C.E. and Dobson, A.P. (2015) Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 23, 172–180.
| Bats as ‘special’ reservoirs for emerging zoonotic pathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXltVehug%3D%3D&md5=7923a438e35c409ff42faf171940ee32CAS |
[3] Luis, A.D. et al. (2015) Network analysis of host–virus communities in bats and rodents reveals determinants of cross‐species transmission. Ecol. Lett. 18, 1153–1162.
| Network analysis of host–virus communities in bats and rodents reveals determinants of cross‐species transmission.Crossref | GoogleScholarGoogle Scholar |
[4] Han, B.A. et al. (2016) Global patterns of zoonotic disease in mammals. Trends Parasitol. 32, 565–577.
| Global patterns of zoonotic disease in mammals.Crossref | GoogleScholarGoogle Scholar |
[5] Romanelli, C. et al. (2015) Connecting global priorities: biodiversity and human health: a state of knowledge review. World Health Organization/Secretariat of the UN Convention on Biological Diversity.
[6] Restif, O. et al. (2012) Model‐guided fieldwork: practical guidelines for multidisciplinary research on wildlife ecological and epidemiological dynamics. Ecol. Lett. 15, 1083–1094.
| Model‐guided fieldwork: practical guidelines for multidisciplinary research on wildlife ecological and epidemiological dynamics.Crossref | GoogleScholarGoogle Scholar |
[7] Wood, J.L. et al. (2012) A framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2881–2892.
[8] Plowright, R.K. et al. (2015) Ecological dynamics of emerging bat virus spillover. Proc. R. Soc. Lond. B Biol. Sci. 282, 20142124.
[9] Streicker D.G. et al 2016 Host–pathogen evolutionary signatures reveal dynamics and future invasions of vampire bat rabies. Proc. Natl. Acad. Sci. USA 201606587
[10] Lloyd-Smith, J.O. et al. (2009) Epidemic dynamics at the human-animal interface. Science 326, 1362–1367.
| Epidemic dynamics at the human-animal interface.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsV2gsLjF&md5=3306671156d6cd4c5d1786565a102a04CAS |
[11] O’Shea, T.J. et al. (2016) Multiple mortality events in bats: a global review. Mammal Rev. 46, 175–190.
| Multiple mortality events in bats: a global review.Crossref | GoogleScholarGoogle Scholar |
[12] Murray, K. et al. (1995) A novel morbillivirus pneumonia of horses and its transmission to humans. Emerg. Infect. Dis. 1, 31–33.
| A novel morbillivirus pneumonia of horses and its transmission to humans.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2s%2FmsVShsQ%3D%3D&md5=92358e5f29eccd3bf5940f6719b2795dCAS |
[13] Philbey, A.W. et al. (1998) An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 4, 269–271.
| An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK1c3osVaitQ%3D%3D&md5=e24100b0447a9b5f3be0694147647abdCAS |
[14] Middleton, D. et al. (2014) Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis. 20, 372–379.
| Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health.Crossref | GoogleScholarGoogle Scholar |
[15] Pulliam, J.R. et al. (2011) Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J. R. Soc. Interface 9, 89–101.
| Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis.Crossref | GoogleScholarGoogle Scholar |
[16] Gurley, E.S. et al. (2007) Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 13, 1031–1037.
| Person-to-person transmission of Nipah virus in a Bangladeshi community.Crossref | GoogleScholarGoogle Scholar |
[17] WHO Ebola Response Team et al. (2015) West African Ebola epidemic after one year—slowing but not yet under control. N. Engl. J. Med. 372, 584–587.
| West African Ebola epidemic after one year—slowing but not yet under control.Crossref | GoogleScholarGoogle Scholar |
[18] Plowright, R.K. et al. (2011) Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. R. Soc. 278, 3703–3712.
| Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.).Crossref | GoogleScholarGoogle Scholar |
[19] Giles, J.R. et al. (2016) Models of Eucalypt phenology predict bat population flux. Ecol. Evol. 6, 7230–7245.
| Models of Eucalypt phenology predict bat population flux.Crossref | GoogleScholarGoogle Scholar |
[20] Hahn, M.B. et al. (2014) The role of landscape composition and configuration on Pteropus giganteus roosting ecology and Nipah virus spillover risk in Bangladesh. Am. J. Trop. Med. Hyg. 90, 247–255.
| The role of landscape composition and configuration on Pteropus giganteus roosting ecology and Nipah virus spillover risk in Bangladesh.Crossref | GoogleScholarGoogle Scholar |
[21] Rulli, M.C. et al. (2017) Is there a nexus between deforestation in Africa and Ebola virus disease outbreaks? Sci. Rep. , .
[22] Plowright, R.K. et al. (2008) Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc. Biol. Sci. 275, 861–869.
[23] Zhou, P. et al. (2016) Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. USA 113, 2696–2701.
| Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XivVKjs74%3D&md5=a7ea507734793e042d456255e30440dbCAS |
[24] Martin, G.A. et al. (2016) Climatic suitability influences species specific abundance patterns of Australian flying foxes and risk of Hendra virus spillover. One Health 2, 115–121.
| Climatic suitability influences species specific abundance patterns of Australian flying foxes and risk of Hendra virus spillover.Crossref | GoogleScholarGoogle Scholar |
[25] Martin, G. et al. (2015) Hendra virus survival does not explain spillover patterns and implicates relatively direct transmission routes from flying foxes to horses. J. Gen. Virol. 96, 1229–1237.
| Hendra virus survival does not explain spillover patterns and implicates relatively direct transmission routes from flying foxes to horses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXht12isb%2FE&md5=9b264221520cc712dea8a71881bd0595CAS |
Biographies
Dr David A Wilkinson studied Molecular and Cellular Biochemistry at University of Oxford, UK obtaining both MBiochem and DPhil degrees. He has subsequently used these skills to study zoonotic infectious diseases. He utilises a combination of classical microbiology, genetics and state-of-the-art genomics to study the evolution and epidemiology of disease-causing microorganisms. After several years studying endemic and emerging bacteria and viruses, including from bats, for Centre de Recherche et de Veille sur les Maladies Émergentes dans l’Océan Indien, France, he was awarded a postdoctoral fellowship at the Molecular Epidemiology and Public Health Laboratory (mEpiLab), Massey University, New Zealand. In New Zealand he studies a number of systems including Campylobacter and Leptospira in domestic and wild animals.
Dr David Hayman is an Associate Professor in Veterinary Public Health and co-directs the Molecular Epidemiology and Public Health Laboratory (mEpiLab) at Massey University, New Zealand. The mEpiLab develops and applies new techniques to inform decision making and guide the prevention and control of infectious disease and forms part of an OIE collaborating Centre for Veterinary Epidemiology and Public Health. Previous positions include a David H Smith Conservation Postdoctoral Fellowship at Colorado State University and University of Florida, USA; a Wellcome Trust Research Training Fellowship and Cambridge Infectious Diseases Consortium Fellowship at University of Cambridge, UK, from where he obtained PhD. He has worked in the Wildlife Zoonoses and Vector-borne Diseases Group at the Animal and Plant Health Agency and Institute of Zoology, and holds a veterinary degree from Edinburgh and a Conservation Biology MSc from Kent, UK.


