Vaccination against streptococcal infections in farmed fish
Andrew C Barnes A B and Oleksandra Rudenko AA The University of Queensland School of Biological Sciences and Centre for Marine Science Brisbane, Qld 4072, Australia
B Email: a.barnes@uq.edu.au
Microbiology Australia 37(3) 118-121 https://doi.org/10.1071/MA16040
Published: 10 August 2016
Aquaculture produces more than 50% of fish for human consumption and, in spite of major improvements since the adoption of injectable vaccines in the 1990s, bacterial diseases still account for considerable losses, particularly in tropical and warm temperate species. Streptococcosis, caused predominantly by Streptococcus iniae and S. agalactiae, manifests as a generalised septicaemia and meningitis followed by rapid mortality. Vaccination against streptococcal infections is difficult as a result of multiple, poorly defined serotypes and consequent vaccine escape (reinfection of previously vaccinated animals). However, genomics applied to reverse vaccinology is providing novel insights into diversity among these aquatic pathogens and is identifying cross-serotype targets that may be exploited for new generation streptococcal vaccines for aquaculture.
Aquaculture reached a significant milestone in 2013 as global production for food use overtook beef production for the first time and now accounts for more than 50% of the global seafood supply1. Aquaculture has wrestled with social license throughout its rapid growth in developed economies, including Australia, through the 1980s and 90s2,3. Objections have been raised around environmental issues such as eutrophication of marine sediments, escape of domesticated fish into wild stocks and pressure on wild fisheries for fishmeal for aquaculture diets. Disease transmission between farmed and wild stock and high antibiotic use for controlling bacterial infections in farmed fish have also attracted attention. Granted, antibiotic use was high in salmonid aquaculture in the 1980s and early 90s, with aquaculture outstripping both human and terrestrial animal use in Norway4. However, the widespread adoption of oil-adjuvanted injectable vaccines in salmonid aquaculture during the mid-1990s all but eliminated antibiotic use from salmonid production4,5. Nevertheless, most current and future expansion of finfish aquaculture is occurring in warm temperate and tropical regions where farmed species and the diseases from which they are at risk are not yet adequately controlled by vaccination.
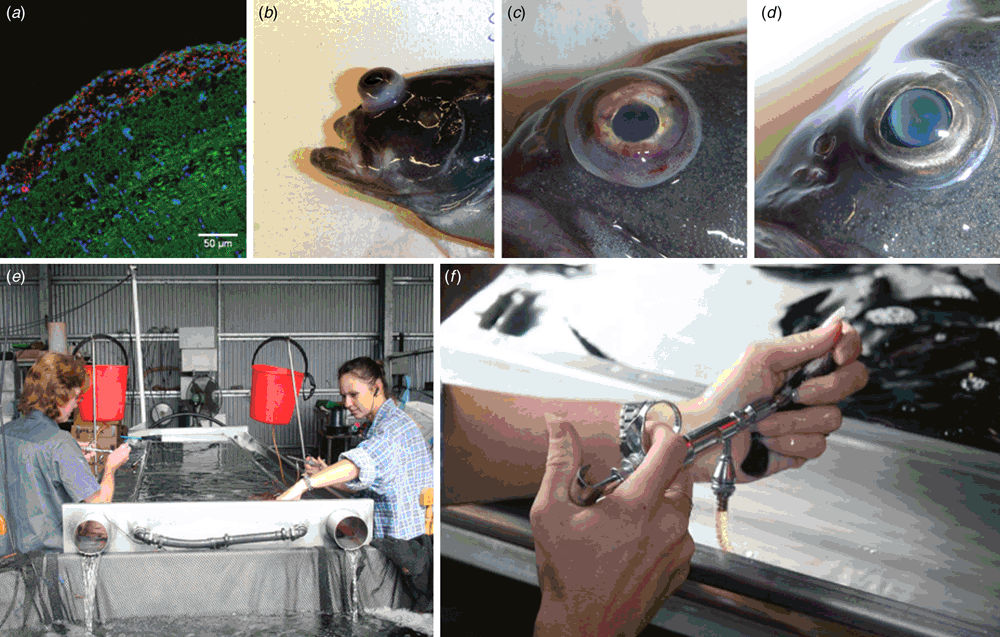
Streptococcal infections occur in warm-temperate and tropical waters wherever fish are farmed and occasionally cause wild fish kills6. While there are a number of streptococcal species that cause disease in fish, the most prevalent and damaging are S. iniae and S. agalactiae. Infection of fish by either pathogen results in rapid onset of generalised septicaemia, meningitis often associated with bilateral exophthalmia (Figure 1a–d) and death with mortalities often exceeding more than 70% within a few days of infection in experimental models.
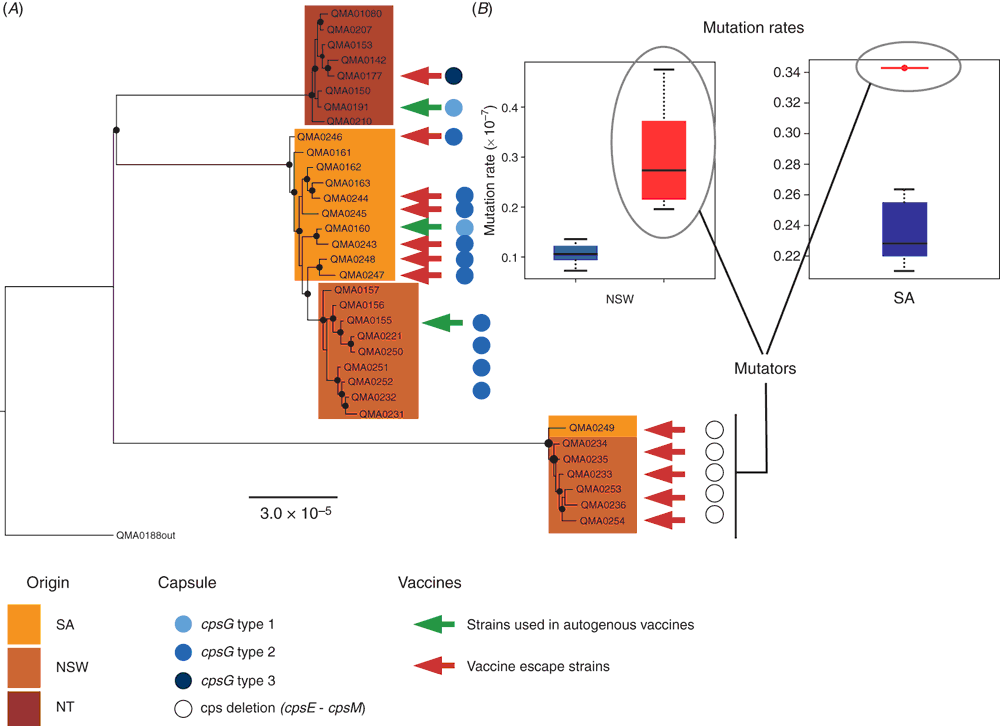
Streptococcus iniae was first isolated in 1972 from an abscess on a captive Amazon freshwater dolphin Inia geoffrensis from which it derives its name7. In aquaculture, the major species affected by S. iniae are rainbow trout in Israel8, grouper in Taiwan9, tilapia, catfish and hybrid bass in the USA10 and, in Australia, barramundi11. Vaccination against S. iniae is accomplished using formalin inactivated bacterins by intraperitoneal injection of fish under general anaesthetic in tilapia12, trout13, grouper9 and barramundi (Figure 1e, f)14. But such vaccines are serotype-specific14–16 and in the absence of a robust serotyping scheme or typing antisera, vaccine failures may occur13,14 where the prevalent strain does not match the serotype of the vaccine used. Serotype is defined by the polysaccharide capsule in S. iniae8,14,17,18 and antigenic changes result from non-synonymous mutations in a limited repertoire of the genes in the capsular operon that alter monomer composition, polymer chain length and quantity of the capsule14. When a vaccine is deployed, new serotypes periodically arise through mutations in variable capsular genes. These serotypes may be already present in the pool of extant strains co-existing on the farm (serotype replacement, adaptation through standing variation) or originate under immune pressure (adaptation through de novo variation). Whole genome sequencing coupled with fluctuation analysis (a statistical method of measuring mutation rate in bacteria based on frequency with which resistance to antibiotic occurs in highly replicated laboratory experiments) suggests both are likely to occur, with a role for hypermutators (variants with greatly elevated mutation rates) facilitating adaptation to the immune host (Figure 2). To overcome the inherent plasticity in the capsular structure, several proteins have been proposed as potential vaccine immunogens based on analysis of the first available genome sequences21, but these have not been uniformly effective22–24. With the falling cost of whole genome sequencing, using informatics to identify surface associated and secreted proteins that are conserved across different capsular serotypes has become a fast and cost-effective route to new experimental vaccines that may be cross-protective across multiple serotypes.
|
Streptococcus agalactiae is a Lancefield group B Streptococcus (GBS). Most fish-pathogenic isolates fall into either (multilocus sequence type) ST260 or ST261 with capsular serotypes Ia and Ib. There have also been reports of fish disease caused by the broad host range ST7 but these have been associated with environmental contamination from terrestrial sources25,26. The ‘true’ fish pathogens are quite distinct from their terrestrial con-specifics, with substantially reduced genomes, depleted virulence factor repertoire and reduction of carbohydrate metabolic pathway genes26. Indeed, fish pathogenic GBS was classified as a separate species, S. ‘difficilis’, until these isolates were later assigned to the species S. agalactiae based on whole cell protein analysis in the late 1990s27 and confirmed by DNA : DNA hybridisation in 200528. Infection and mortality caused by GBS is one of the most significant issues facing tilapia culture globally. Injectable vaccines are effective but type-specific29. Once again the potential for exploiting the falling costs of whole genome sequencing for design of cross-serotype protective vaccines is substantial and has been used to great effect in human medicine30. A similar approach is ongoing for aquatic isolates, and surface expressed proteins unique to aquatic isolates but conserved across the ST260 and ST261 sequence types have been identified and tested for efficacy in preliminary trials in tilapia.
Whilst large-scale whole genome sequencing is identifying antigens conserved across the most important serotypes, there are a number of further problems that must be resolved for viable streptococcal vaccines for aquaculture. First, fish are a low value commodity and even in salmon, which fetch a relatively high wholesale price, margin per dose of vaccine is low relative to other animal vaccines, except poultry. Consider that the farm gate price of most warm water species is substantially lower than salmon, with tilapia valued at less than one third of the lowest salmon price, and one can envisage that the cost per dose of vaccine has to be very low indeed. Whilst there is some margin in simple formalin-killed bacterins, recombinant protein vaccines are not economically viable in this market. Therefore, maximising expression of conserved antigens, identified through genomics, in culture for improvement of killed bacterins makes more commercial sense. The second problem relates to adjuvants. The success of vaccination in cool water salmonid aquaculture was founded upon oil emulsions that enable a single vaccination to protect for the complete farm lifecycle. This duration of immunity in excess of two years necessitates very slow antigen release from the emulsion. This works against warm water species that are farmed for maybe 9–12 months in the case of tilapias, particularly for streptococcal vaccines, where achieving an effective antigen dose against non-carbohydrate antigens is already challenging due to very low growth densities of aquatic streptococci. This will necessitate clever formulation of vaccines to enable initial fast antigen release, but also sustained protection for several months, and all at a price of a few cents per dose. This represents a substantial challenge for the industry and the science.
References
[1] Anon. (2014) The State of World Fisheries and Aquaculture. In: FAO, ed. Food and Agriculture Organisation of the United Nations, Rome.[2] Hamouda, L. et al. (2005) The salmon aquaculture conflict in British Columbia: a graph model analysis. Ocean Coast. Manage. 48, 571–587.
| The salmon aquaculture conflict in British Columbia: a graph model analysis.Crossref | GoogleScholarGoogle Scholar |
[3] Leith, P. et al. (2014) Science and social license: defining environmental sustainability of Atlantic salmon aquaculture in south-eastern Tasmania, Australia. Soc. Epistemol. 28, 277–296.
| Science and social license: defining environmental sustainability of Atlantic salmon aquaculture in south-eastern Tasmania, Australia.Crossref | GoogleScholarGoogle Scholar |
[4] Grave, K. et al. (1999) Surveillance of the overall consumption of antibacterial drugs in humans, domestic animals and farmed fish in Norway in 1992 and 1996. J. Antimicrob. Chemother. 43, 243–252.
| Surveillance of the overall consumption of antibacterial drugs in humans, domestic animals and farmed fish in Norway in 1992 and 1996.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXhsFyms7o%3D&md5=bd0e08edeffafde2dd9b0ea65991f6a6CAS | 11252330PubMed |
[5] Sommerset, I. et al. (2005) Vaccines for fish in aquaculture. Expert Rev. Vaccines 4, 89–101.
| Vaccines for fish in aquaculture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXis12nsro%3D&md5=b509b89be80d911921d4a50fadc77b0eCAS | 15757476PubMed |
[6] Agnew, W. and Barnes, A.C. (2007) Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet. Microbiol. 122, 1–15.
| Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXksFWqtbY%3D&md5=cab24d16e9591ec67b08f865aed94882CAS | 17418985PubMed |
[7] Pier, G.B. and Madin, S.H. (1976) Streptococcus iniae sp nov, a beta-hemolytic Streptococcus isolated from an amazon freshwater dolphin, Inia geoffrensis. Int. J. Syst. Bacteriol. 26, 545–553.
| Streptococcus iniae sp nov, a beta-hemolytic Streptococcus isolated from an amazon freshwater dolphin, Inia geoffrensis.Crossref | GoogleScholarGoogle Scholar |
[8] Lahav, D. et al. (2004) Streptococcus iniae type II infections in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 62, 177–180.
| Streptococcus iniae type II infections in rainbow trout Oncorhynchus mykiss.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD2M%2Fit1Ggug%3D%3D&md5=dff168db68c71a2d67c7a7ccd46ed99fCAS | 15648844PubMed |
[9] Huang, H.Y. et al. (2014) Efficacy of a formalin-inactivated vaccine against Streptococcus iniae infection in the farmed grouper Epinephelus coioides by intraperitoneal immunization. Vaccine 32, 7014–7020.
| Efficacy of a formalin-inactivated vaccine against Streptococcus iniae infection in the farmed grouper Epinephelus coioides by intraperitoneal immunization.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsVymtr%2FK&md5=42b245d597a42108c82eaf403b6fc135CAS | 25192808PubMed |
[10] Shoemaker, C.A. et al. (2001) Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States. Am. J. Vet. Res. 62, 174–177.
| Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD38%2Fgs1amsw%3D%3D&md5=32aaa5592076ce96f133e073da538a19CAS | 11212023PubMed |
[11] Bromage, E.S. et al. (1999) Streptococcus iniae, a bacterial infection in barramundi Lates calcarifer. Dis. Aquat. Organ. 36, 177–181.
| Streptococcus iniae, a bacterial infection in barramundi Lates calcarifer.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK1MzivFyhtA%3D%3D&md5=dc1b868c3bd900fc7b50ce0d6f5e201aCAS | 10401583PubMed |
[12] Klesius, P.H. et al. (1999) Efficacy of a killed Streptococcus iniae vaccine in tilapia (Oreochromis niloticus). Bull. Eur. Assoc. Fish Pathol. 19, 39–41.
[13] Bachrach, G. et al. (2001) Recovery of Streptococcus iniae from diseased fish previously vaccinated with a streptococcus vaccine. Appl. Environ. Microbiol. 67, 3756–3758.
| Recovery of Streptococcus iniae from diseased fish previously vaccinated with a streptococcus vaccine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlvFSksL0%3D&md5=b2a4eaf2a13c1b313cd9eea1ad5327cdCAS | 11472962PubMed |
[14] Millard, C.M. et al. (2012) Evolution of the capsular operon of Streptococcus iniae in response to vaccination. Appl. Environ. Microbiol. 78, 8219–8226.
| Evolution of the capsular operon of Streptococcus iniae in response to vaccination.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1yktr3F&md5=4fe44a493e5c9141ddda728a7d2e20c3CAS | 23001668PubMed |
[15] Klesius, P.H. et al. (2000) Efficacy of single and combined Streptococcus iniae isolate vaccine administered by intraperitoneal and intramuscular routes in tilapia (Oreochromis niloticus). Aquaculture 188, 237–246.
| Efficacy of single and combined Streptococcus iniae isolate vaccine administered by intraperitoneal and intramuscular routes in tilapia (Oreochromis niloticus).Crossref | GoogleScholarGoogle Scholar |
[16] Shoemaker, C.A. et al. (2010) Protection against heterologous Streptococcus iniae isolates using a modified bacterin vaccine in Nile tilapia, Oreochromis niloticus (L.). J. Fish Dis. 33, 537–544.
| Protection against heterologous Streptococcus iniae isolates using a modified bacterin vaccine in Nile tilapia, Oreochromis niloticus (L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXptV2ntLw%3D&md5=797c4085a9129615098a6d59f26495e3CAS | 20298447PubMed |
[17] Eyngor, M. et al. (2008) Emergence of novel Streptococcus iniae exopolysaccharide-producing strains following vaccination with nonproducing strains. Appl. Environ. Microbiol. 74, 6892–6897.
| Emergence of novel Streptococcus iniae exopolysaccharide-producing strains following vaccination with nonproducing strains.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsVaru73N&md5=bb73b38c63498c4bc87badc91fc1ec31CAS | 18806000PubMed |
[18] Barnes, A.C. et al. (2003) Streptococcus iniae: serological differences, presence of capsule and resistance to immune serum killing. Dis. Aquat. Organ. 53, 241–247.
| Streptococcus iniae: serological differences, presence of capsule and resistance to immune serum killing.Crossref | GoogleScholarGoogle Scholar | 12691195PubMed |
[19] Croucher, N.J. et al. (2015) Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15.
| Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins.Crossref | GoogleScholarGoogle Scholar | 25414349PubMed |
[20] Leaché, A.D. et al. (2015) Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Syst. Biol. 64, 1032–1047.
| Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies.Crossref | GoogleScholarGoogle Scholar | 26227865PubMed |
[21] Baiano, J.C. and Barnes, A.C. (2009) Towards control of Streptococcus iniae. Emerg. Infect. Dis. 15, 1891–1896.
| Towards control of Streptococcus iniae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitlajug%3D%3D&md5=009303351e12b90d84501df26675a62fCAS | 19961667PubMed |
[22] Aviles, F. et al. (2013) The conserved surface M-protein SiMA of Streptococcus iniae is not effective as a cross-protective vaccine against differing capsular serotypes in farmed fish. Vet. Microbiol. 162, 151–159.
| The conserved surface M-protein SiMA of Streptococcus iniae is not effective as a cross-protective vaccine against differing capsular serotypes in farmed fish.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlGnsLnL&md5=d6fc00fad713bd2b65d8a3b971921212CAS | 22989514PubMed |
[23] Membrebe, J.D. et al. (2016) Protective efficacy of Streptococcus iniae derived enolase against Streptococcal infection in a zebrafish model. Vet. Immunol. Immunopathol. 170, 25–29.
| Protective efficacy of Streptococcus iniae derived enolase against Streptococcal infection in a zebrafish model.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xhs1Oisbk%3D&md5=bac86b78f3c0c445095be61d070d14baCAS | 26872628PubMed |
[24] Sun, Y. et al. (2013) SagE induces highly effective protective immunity against Streptococcus iniae mainly through an immunogenic domain in the extracellular region. Acta Vet. Scand. 55, 78.
| SagE induces highly effective protective immunity against Streptococcus iniae mainly through an immunogenic domain in the extracellular region.Crossref | GoogleScholarGoogle Scholar | 24215645PubMed |
[25] Jafar, Q.A. et al. (2009) Molecular investigation of Streptococcus agalactiae isolates from environmental samples and fish specimens during a massive fish kill in Kuwait Bay. Afr. J. Microbiol. Res. 3, 22–26.
| 1:CAS:528:DC%2BD1MXhtFaqurfP&md5=9c1d709dd99f449e9ddc806f54353ab6CAS |
[26] Rosinski-Chupin, I. et al. (2013) Reductive evolution in Streptococcus agalactiae and the emergence of a host adapted lineage. BMC Genomics 14, 252.
| Reductive evolution in Streptococcus agalactiae and the emergence of a host adapted lineage.Crossref | GoogleScholarGoogle Scholar | 23586779PubMed |
[27] Vandamme, P. et al. (1997) Streptococcus difficile is a nonhemolytic group B, type Ib Streptococcus. Int. J. Syst. Bacteriol. 47, 81–85.
| Streptococcus difficile is a nonhemolytic group B, type Ib Streptococcus.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2s7kvV2rsA%3D%3D&md5=a2043ff69d5cbd27a22171d37b42f9d4CAS | 8995807PubMed |
[28] Kawamura, Y. et al. (2005) High genetic similarity of Streptococcus agalactiae and Streptococcus difficilis: S. difficilis Eldar et al. 1995 is a later synonym of S-agalactiae Lehmann and Neumann 1896 (Approved Lists 1980). Int. J. Syst. Evol. Microbiol. 55, 961–965.
| High genetic similarity of Streptococcus agalactiae and Streptococcus difficilis: S. difficilis Eldar et al. 1995 is a later synonym of S-agalactiae Lehmann and Neumann 1896 (Approved Lists 1980).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjsFehsr8%3D&md5=110fadc3212491b17cb803a0803627efCAS | 15774692PubMed |
[29] Evans, J.J. et al. (2004) Efficacy of Streptococcus agalactiae (group B) vaccine in tilapia (Oreochromis niloticus) by intraperitoneal and bath immersion administration. Vaccine 22, 3769–3773.
| Efficacy of Streptococcus agalactiae (group B) vaccine in tilapia (Oreochromis niloticus) by intraperitoneal and bath immersion administration.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmsl2jt74%3D&md5=8707bdab8c5928c51a4cf71cf7dcf575CAS | 15315858PubMed |
[30] Maione, D. et al. (2005) Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309, 148–150.
| Identification of a universal Group B streptococcus vaccine by multiple genome screen.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXlsF2ht7o%3D&md5=5c865d01e3442f427152ea32afeadbfaCAS | 15994562PubMed |
Biographies
Andy Barnes is Associate Professor in Aquatic Animal Health at The University of Queensland, with research activities focusing on preventative healthcare in commercially important aquatic animals including finfish, penaied and freshwater prawns and oysters. After completing a PhD in medical microbiology at The University of Edinburgh Medical School in 1992, Andy joined Aqua Health Limited, market leader in aquaculture vaccines, in research and development of vaccines for fish before its takeover by Novartis Animal Health in 2000. Andy joined The University of Queensland from Novartis in October 2003. Recent research projects include tailoring generic vaccines against Streptococcosis for the global market, and specific vaccines for the specialist Australian market. He recently completed an evolutionary history of Yersinia ruckeri in Tasmania and New Zealand by whole genome analysis. His interests lie in evolution of bacterial pathogens of aquatic animals and adaptation to the immune host.
Oleksandra Rudenko is a PhD student at The University of Queensland, primarily interested in microbial evolution and interactions with a host. After she graduated as MSc in Biology at Uzhhorod National University, department of Genetics, Plant Physiology and Microbiology in 2008, she worked in bacteriological laboratory at Uzhhorod Central City Hospital up until her UQ studies commenced in 2014. Her project focuses on the role of hypermutable strains in microbial adaptation to the host and immune escape after vaccination.