Composition of dissolved organic matter within a lacustrine environment
Margaret V. McCaul A , David Sutton C , André J. Simpson B , Adrian Spence A , David J. McNally B , Brian W. Moran A , Alok Goel D , Brendan O’Connor D , Kris Hart A and Brian P. Kelleher A EA School of Chemical Sciences, Dublin City University, Glasnevin, Dublin 9, Ireland.
B Department of Chemistry, University of Toronto, Scarborough College, 1265 Military Trail, Toronto, ON, M1C1A4, Canada.
C School of Science, Limerick Institute of Technology, Limerick, Ireland.
D Centre for Bioanalytical Sciences and School of Biotechnology, DCU, Glasnevin, Dublin 9, Ireland.
E Corresponding author. Email: brian.kelleher@dcu.ie
Environmental Chemistry 8(2) 146-154 https://doi.org/10.1071/EN10109
Submitted: 4 October 2010 Accepted: 17 December 2010 Published: 2 May 2011
Journal Compilation © CSIRO Publishing 2011 Open Access CC BY-NC-ND
Environmental context. Freshwater dissolved organic matter is a complex chemical mixture central to many environmental processes, including carbon and nitrogen cycling. Questions remain, however, as to its chemical characteristics, sources and transformation mechanisms. We studied the nature of dissolved organic matter in a lake system and found that it is influenced by anthropogenic activities. Human activities can therefore influence the huge amounts of carbon sequestered in lakes as dissolved organic matter.
Abstract. Freshwater dissolved organic matter (DOM) is a complex mixture of chemical components that are central to many environmental processes, including carbon and nitrogen cycling. However, questions remain as to its chemical characteristics, sources and transformation mechanisms. Here, we employ 1- and 2-D nuclear magnetic resonance (NMR) spectroscopy to investigate the structural components of lacustrine DOM from Ireland, and how it varies within a lake system, as well as to assess potential sources. Major components found, such as carboxyl-rich alicyclic molecules (CRAM) are consistent with those recently identified in marine and freshwater DOM. Lignin-type markers and protein/peptides were identified and vary spatially. Phenylalanine was detected in lake areas influenced by agriculture, whereas it is not detectable where zebra mussels are prominent. The presence of peptidoglycan, lipoproteins, large polymeric carbohydrates and proteinaceous material supports the substantial contribution of material derived from microorganisms. Evidence is provided that peptidoglycan and silicate species may in part originate from soil microbes.
Additional keywords: carboxyl-rich alicyclic molecules, NMR, peptidoglycan.
Introduction
Dissolved organic matter (DOM), both marine and freshwater, comprises the largest pool of exchangeable carbon on the Earth’s surface and is derived from numerous sources that influence its relative reactivity and our ability to predict its storage capacity and turnover times.[1,2] Terrestrial and freshwater DOM, whose input to ocean waters is largely controlled by riverine sources, experiences an annual flux of ~0.4 ×1015 g C year–1 to the marine environment.[3] The cycling of DOM from fresh to marine water is not only important in the global carbon cycle but also plays an important role in the enhanced solubility, bioavailability and fate of chemical contaminants and their global transport.[4,5]
Despite this importance, there is still much to learn about the chemical composition of freshwater DOM and how chemical constituents vary worldwide, and between freshwater and marine environments.[6,7] The application of NMR spectroscopy to study structures and interactions in environmental chemistry is growing and is a powerful tool in helping unravel the key structural components in major global carbon pools.[8–11] In recent work, 1- and 2-D solution state NMR spectroscopy has shown that major structural components of lake freshwater include carboxyl-rich alicyclic molecules (CRAM), heteropolysaccharides and aromatic compounds.[12] These components were first reported, and are consistent with those identified, in marine DOM.[13] Furthermore, it has been tentatively suggested that CRAM may be derived from cyclic terpenoids.[12,13] However, it is not clear whether these precursors are of terrestrial or aquatic origin or whether transformations proceed via biological and photochemical processes.
Traditional methods of DOM isolation require large sample volumes to overcome the low concentration in natural waters[14,15] or are laborious and time consuming.[16] Sampling is often carried out over just 1 or 2 days, which is unlikely to be long enough to provide a representative sample of the area. The samplers employed in this study were deployed over a 4-week period and provide a more representative material that is less susceptible to specific daily fluxes. Another advantage of using passive samplers of this kind is that filtration is not required, reducing the possibility of contamination and loss of material. It has also been shown that the material collected on the samplers is similar to that collected using conventional DEAE-cellulose batch extraction, indicating that the passive sampler approach isolates the same components.[17] The same study also reported that 72–89% of total DOM can be captured on the sampler, with the majority of lost material comprising low molecular weight sugars.
Although recent studies have contributed greatly to our knowledge of the overall composition of DOM, less is known of its mechanisms of formation, compositional variation and the origin of the most refractory DOM. Here we use DEAE-cellulose passive samplers, as reported by Lam and Simpson,[17] to concentrate DOM from different areas in Lough Derg, a large lake system on the River Shannon in Ireland. The River Shannon is the largest catchment within Ireland and Britain, draining a land area of ~18 000 km2. Lough Derg, the third largest lake in Ireland, is located at the southern end of the Shannon and covers an area of 120 km2. NMR spectroscopy (both 1- and 2-D) is employed to study DOM structure and how it varies within a lake system and assess anthropogenic influence on its composition. The potential of surrounding soil microbial biomass as a source of DOM is also investigated by comparison of the NMR spectra of degraded soil microbial biomass and leachate to the DOM spectra.
Materials and methods
Sampling and sample preparation
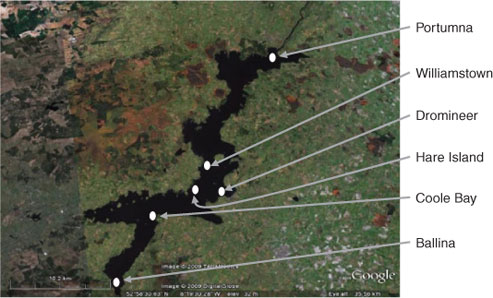
Six sampling sites around Lough Derg (Fig. 1) were chosen to represent areas influenced by different aspects of the surrounding landscape. At each site, two passive samplers (containing six membranes) were placed and suspended (using a fishing line) ~100 cm below the surface of the water. Samples were removed from the lake after 28 days. Sampling at the same sites was carried out in August 2008 and January 2009, so as to assess temporal variation in DOM components. DOM was isolated using a passive sampler.[17] Water from the lough was prefiltered through 0.22-μm poly(vinylidene difluoride) (PVDF) filters (Spectrapor). DOM was isolated on diethylaminoethyl cellulose resin (Sigma Aldrich), a selective resin that adsorbs negatively charged species at neutral pH. The cellulose resin is contained within the PVDF tubing and protected via a high density polyethylene (HDPE) casing with predrilled holes. Prior to use, DEAE-cellulose was precleaned using a cycle of acid, base and distilled water washings. Cleaned DEAE-cellulose (250 mg) was slurry packed with distilled water into 7 cm long (24 mm wide) PVDF porous membranes, which were pre-soaked in 0.1% sodium azide for a minimum of 48 h.
|
Extraction of bound DOM from the passive samplers was performed by cutting and removing the resin from the PVDF membranes. The resin was then placed in 50-mL Teflon centrifuge tubes and extracted using ~40 mL of 0.1-M NaOH. The tubes were centrifuged (10 000g, 10 min at 23°C) to pellet the resin, and the supernatant was decanted. The pellet was re-suspended and the previous steps were repeated four times, or until the extracting solvent was colourless, to ensure complete extraction of DOM from the resin. The extracted DOM was ion-exchanged using Amberjet 1200H Plus resin (Aldrich) and freeze-dried. Duplicate samples were freeze-dried and samples were re-suspended in deuterium oxide (D2O) for NMR spectroscopic analysis.
NMR spectroscopic analysis
Each sample (100 mg) was dissolved in 1 mL of D2O and titrated to pH 13.1 using NaOD (40% by wt) to ensure complete solubility. Samples were analysed using a Bruker Avance 500 MHz NMR spectrometer equipped with a 1H–BB–13C 5 mm, triple resonance broadband inverse probe at 298 K. 1-D solution state 1H NMR experiments were performed with 256 scans, a recycle delay of 3 s, 32 768 time domain points, and an acquisition time of 1.6 s. Solvent suppression was achieved by presaturation utilising relaxation gradients and echoes.[18] Spectra were apodised through multiplication with an exponential decay corresponding to 1-Hz line broadening, and a zero-filling factor of 2. Diffusion-edited (DE) experiments were performed using a bipolar pulse longitudinal encode-decode sequence.[19] Scans (1024) were collected using a 2.5-ms, 49-gauss cm–1, sine-shaped gradient pulse, a diffusion time of 100 ms, 8192 time domain points and 410 ms acquisition time. Spectra were apodised through multiplication with an exponential decay corresponding to 10 Hz line broadening and zero-filling factor of 2.
Total correlation spectroscopy (TOCSY) spectra were obtained in the phase sensitive mode, using time proportional phase incrimination (TPPI). TOCSY with presaturation of the solvent resonance was acquired using 2048 time domain points in the F2 dimension and 128 scans for each of the 128 slices in the F1 dimension. A mixing time of 60 ms was used with a relaxation delay of 1 s. Processing of both dimensions used a sine-squared function with a π/2 phase shift and a zero-filling factor of 2. TOCSY data was collected to help confirm the major assignments highlighted on the 1H–13C NMR correlations.
Heteronuclear multiple quantum coherence (HMQC) spectra were obtained in phase sensitive mode using Echo/Antiecho gradient selection. The HMQC experiments were carried out using 256 scans with 128 time domain points in the F1 dimension and 1024 time domain points in the F2 dimension. A relaxation delay of 1 s and 1J 1H–13C of 145 Hz were used. F2 dimensions in HMQC experiments were processed using an exponential function corresponding to a 15 Hz line broadening. The F1 dimension was processed using a sine-squared function with a π/2 phase shift and a zero-filling factor of 2.
Spectral predictions were carried out using Advanced Chemistry Development’s ACD/SpecManager and ACD/2-D NMR Predictor using Neural Network Prediction algorithms (version 10.02). Parameters used for prediction including line shape, spectral resolution, sweep width and spectrometer frequency were set to match those of the real datasets as closely as possible. Please see the Accessory publication for an example (http://www.publish.csiro.au/?act=view_file&file_id=EN10109_AC.pdf).
Growth and degradation of soil microbial biomass
The soil used in this study is a light clay loam from a cultivated field near Lough Derg. Sampling was carried out according to a modified version of the protocol described processing Joseph et al.[20] A 25-mm diameter clean metal core was used to sample 100 mm-long soil cores from the A horizon, which were transferred to sterile polyethylene bags and sealed at the collection site. Soil cores were transported at the ambient temperature and processed within 24 h of collection. The upper 30 mm of each core was discarded, and large pieces of roots and stones were removed from the remainder, which was sieved through a stainless steel sieve with a 2-mm aperture (IMPACT Laboratory Test Sieve, UK). Sieved samples were pooled, homogenised and stored at 4°C at its field moisture content for further analysis. A CHN combustion analyser (Exeter Analytical CE440 elemental analyser) was used to determine the soil elemental composition, 4.25% C, 0.58% H, 0.15% N and 0.21% P.
Soil microbes were cultivated according to a modified version of the protocol described by Janssen et al.[21] Soil (1 g) was added to 100-mL aliquots of sterile distilled water and dispersed with a magnetic stirrer. Aliquots (1 mL) of soil suspension were added to 9-mL portions of dilute nutrient broth (DNB), containing: Laboratory-Lemco’ powder (1.0 g L–1); yeast extract (2.0 g L–1); peptone (5.0 g L–1); and NaCl (5.0 g L–1), in distilled water (Oxoid Ltd, Hampshire, England). Diluted soil suspensions were mixed by vortexing at ~150 rpm for 10 s and used to prepare serial dilutions containing 10–2 to 10–4 g soil suspension. Aliquots (100 µL) of each dilution series was plated on duplicate LB agar plates containing 0.5% dripstone, 0.25% yeast extract, 0.1% D-glucose, 0.25% NaCl and 1.5% agar. Serially inoculated LB plates were incubated at room temperature for 2 days and all isolated colonies were selected from the 10–4 dilution of the soil and used to inoculate 3.0 mL of LB broth. Cultures were incubated at for 48 h.
The degradation experiment was conducted according to a modified version of the protocol described by Kelleher et al.[22] The experimental design attempted to mimic in situ conditions and enable collection of transformed and leached organic matter (OM) for further analysis. Glass funnels with borosilicate sintered discs, with porosity grade 4 were submerged until flush with soil in a clay pot. The soil used was a native light clay-loam taken from fields surrounding Lough Derg. The cavity beneath the sintered disc was filled with the native soil and secured with glass wool and 0.4 g of the soil microbial biomass evenly distributed on the surface of the sintered disc. This set up enables microbes in the soil to access the microbial biomass. The biomass was sprinkled with water every second day to mimic rain and the runoff was collected in a vial attached to the end of the funnel. Moisture levels were kept constant throughout the experiment. Runoff and microbial biomass were collected at 6 and 14 weeks post-degradation.
Results and discussion
General characterisation
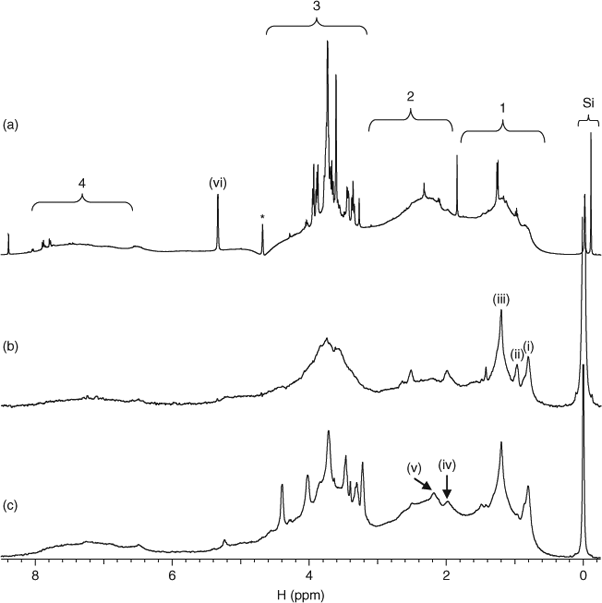
Recent studies that have employed multidimensional NMR spectroscopy to study DOM show that marine and freshwater DOM share many structural similarities.[12,13] These major structural components are also present in all the DOM isolated from Lough Derg. For example, Fig. 2 shows the conventional 1H (Fig. 2a) and diffusion edited (Fig. 2b) NMR spectra for the Ballina DOM sample and also shows the diffusion edited 1H spectrum of the Coole Bay sample area of lake (Fig. 2c).
|
General assignments, consistent with those reported are: (1) aliphatics, including material derived from linear terpanoids; (2) carboxyl-rich alicyclic molecules (CRAM; see also Fig. 5); (3) a mixture of carbohydrates and amino acids; (4) aromatics, including resonances from amino acid (AA) side chains.[12,13] More specific assignments refer to (i) CH3, likely including resonances from aliphatic species and methylated amino acid side-chain residues in peptides/protein; (ii) consistent with a side chain residue also seen in the 1H NMR spectrum for bovine serum albumin; (iii) aliphatic methylene (CH2)n; (iv) contributions from both the N-acetyl group in peptidoglycans and other units lipids and waxes[23,24]; (v) mainly aliphatic methylene units β to an acid or ester i.e. R2-OCO-CH2-R1 or double bond; and (vi) anomeric protons in carbohydrate. ‘Si’ indicates a natural silicate species and not TMS (tetramethylsilane, Si(CH3)4), a commonly used NMR reference standard).[23]
Fig. 2a displays sharp peaks, especially in the carbohydrate region (3). Sharper lines observed in NMR spectra are often characteristic of smaller structures,[22] and this may indicate the breakdown of the carbohydrates from large polymeric structures into smaller fragments. To test this, diffusion edited (DE) NMR spectroscopy was performed on the Ballina sample. In diffusion edited NMR experiments, small molecules are essentially gated from the final spectrum but signals from macromolecules that display little translational diffusion are not gated and appear in the spectrum.[19,25] The diffusion edited spectrum of Ballina DOM is shown in Fig. 2b. Aliphatic chains are prominent, indicating that they have restricted diffusion, which suggests that they may be present in rigid domains or macromolecular structures. The relative intensity of the carbohydrate signals is much less in the diffusion edited spectrum v. the conventional 1H NMR spectrum, suggesting that a large fraction of the carbohydrates in the DOM is present as relatively small mobile entities. However, there is still a considerable contribution from carbohydrate signals in the diffusion edited spectrum, supporting a second fraction of carbohydrate with greater molecular (or aggregate) size.
A characteristic resonance for CH3 in methylated AA side chain residues (Fig. 2, signal i) is easily distinguishable in the diffusion edited NMR spectrum, suggesting the presence of protein/peptide.[23] Furthermore, the resonance at ~1 ppm (Fig. 2, signal ii) is likely attributed to protein/peptide as this peak is also present in the 1H NMR spectrum of bovine serum albumin.[26] Complimentary evidence for protein/peptide presence is provided by the emergence of α protons from AAs in Fig. 3. Proteinaceous compounds are viewed as labile in the environment[27] and their survival and occurrence have been explained through protection mechanisms such as encapsulation and formation of microbially resistant complexes with carbohydrates and lignin.[28–30] Lam et al.[12] detected weak protein/peptide contributions, which were considered to be only a minor component in Lake Ontario DOM. However, the spectra generated indicate that the protein/peptide contribution may vary considerably between DOM from different sources in freshwater environments. It is estimated that plants often contain only 1–5% protein by weight and that protein structures are known to degrade rapidly in a soil environment.[31,32] It seems unlikely that the preservation of plant-derived peptide/protein structures can completely account for the contributions of proteins and peptides in DOM. It is therefore possible that a significant portion of peptide/protein in DOM arises from the cells of dead and living microbes of either aquatic or terrestrial origin.

|
Alternatively, microbially resistant ligno-protein complexes may also account for some of the protein present.[33] Lignin-type signatures were not found in the study of Lake Ontario DOM,[12] but the possibility of lignin contributions to Lough Derg DOM is highlighted by cross peaks that may represent lignin derived O-CH3 units (Fig. 3), often the most intense signal in soil OM.[9,22,34] Methoxy cross-peaks are clearly present in all the lake samples (overlapped with carbohydrate cross-peaks), especially Hare Island and Dromineer. Lignin is a strong indicator of terrestrial plant inputs and may be an indication of the age of DOM and the influence of the surrounding environment. Proteins originating from microbial cells may be encapsulated by, or sorbed to, lignin, making them less susceptible to degradation.
All Lough Derg DOM samples contain a contribution from carbohydrates that are not removed during diffusion editing (Fig. 2b,c) indicating that there is a polymeric carbohydrate component present that could potentially be associated with the cell walls of microorganisms.[12] Signals (iii) and (v) in Fig. 2b,c are consistent with aliphatic structures. The aliphatic (CH2)n peak is dominant, indicating the presence of stable waxes and lipids.[35] Waxes and cutins derived from plants have been identified in abundance in humic extracts,[25] and are likely to be preserved because of their cross-linked structure and hydrophobicity.[36] Fig. 2c shows the DE 1H NMR spectrum for the Coole Bay DOM sample. Signal (v) is particularly prominent and shows similarities to signals from lipoproteins observed in other natural samples.[23] Lipoprotein is a key component of bacterial cells (also plant, animal, yeast, fungal, algal and insect cells), is structurally diverse and is released during bacterial growth,[37] so its presence corroborates the importance of terrestrial microbes as sources of DOM. Microbial contributions are also supported by the presence of signal (iv) in Fig. 2c. This is consistent with peptidoglycan, which comprises up to 90% by weight of Gram-positive bacteria and is the key structural component in all microbial cell walls. That peptidoglycan was found to accumulate is not unexpected as it is resistant (as are microbe cell walls) to many chemical and biological processes and has been found in the most refractory components of soil OM.[23]
Soil microbial contribution
Despite strong microbial signatures in Fig. 2, it is difficult to know from where the microbial residue originates. It has recently been shown that microbial presence in soil far exceeds presently accepted values and that considering the amounts of fresh cellular material in soil extracts, it is probable that the contributions of microorganisms in the terrestrial environment are seriously underestimated.[26] Therefore, soil microbial biomass may also be an important source of freshwater DOM. The potential contribution of surrounding soil microbial biomass to Lough Derg DOM was studied by conducting a complementary laboratory experiment that monitored the degradation of soil microbial biomass cultured from soil sampled near the lake. Degradation occurred over 14 weeks, allowing NMR experiments to be conducted on degraded soil microbial biomass residue and leachate. Fig. 4 compares the DE 1H NMR spectra of the 14-week leachate from degraded soil microbial biomass (A), to the ‘Dromineer’ DOM sample from Lough Derg (B). Characteristic resonances, such as CH3 in methylated AA side chain residues (signal i) and aliphatic methylene (CH2)n (signal ii) that are present in the Dromineer DOM (Fig. 4b) are also present in the microbial leachate. These signals also persist in degraded plant matter, so it is not possible to say that they originate solely from soil microbial biomass.[27] However, peptidoglycan (PG, Fig. 4a,b) is present in both the DOM and the soil microbial biomass leachate, and this is confirmed in the HMQC spectra in Fig. 5. It should be noted that the slight shift in the proton axes of the PG microbial biomass is from the solvent (DMSO) used to swell the microbial biomass for analysis using HR-MAS (High Resolution Magic Angle Spinning) NMR spectroscopy. The presence of peptidoglycan would suggest that complex biomaterials such as those from the cell walls of soil microorganisms can persist in the water environment and that it is possible that the peptidoglycan we see in DOM originally derived from microbes in soil. However, as peptidoglycan may also be produced by aquatic microbes it is not possible to definitively state the source of this material.

|

|
Interestingly, natural silicate species (Si) present in DOM samples are also present in the microbial leachate spectrum. Carbon sequestration in the oceans is known to be coupled with the global cycle of silicon.[38–40] Rivers provide the conduit for ~5 Tmol Si year–1 to the oceans, which is 80% of the total annual flux.[38,41] The remaining 20% comes from dust and submarine hydrothermal sources. It is thought that the ultimate source of continental silicon flux to the oceans is weathering processes in terrestrial biogeosystems.[42,43] However, Sommer et al. have pointed out that silicon dynamics in terrestrial biogeosystems cannot be understood solely by way of mineral weathering.[44]
The silicate species in the NMR spectra are unusual and arise at ~0 ppm (also present in HMQC data) and suggest methylated silica.[45] It is important to note that these signals are not from TMS, the commonly used internal standard for NMR spectroscopy. TMS is insoluble in water and no internal standards (of any kind) were used. Furthermore, similar signals are seen in all the natural water samples that have been analysed directly with NMR spectroscopy. In direct NMR spectroscopy, the water sample is studied ‘as is’, with no pre-concentration or pre-treatment of any type, indicating that these signals must be of natural origin.[46] Silicate species in the soil microbial leachate would therefore suggest that soil microorganisms accumulate their own stable silicon pools and may play a larger role in silicon cycling than presently thought.
DOM variability
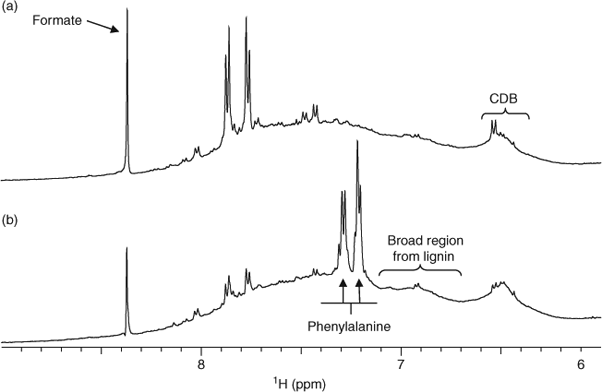
The spectra of samples from different sampling points in the lake that have been presented earlier have been generally similar in composition, although the presence of components such as proteins differ between samples. However, there are smaller differences between samples that may reflect how the surrounding terrestrial environment affects the distribution of DOM within the lacustrine environment. Fig. 6 displays the 1H NMR spectra for the aromatic region of two sample sites in Lough Derg (Coole Bay and Dromineer). The samples display generally similar profiles and ratios of major chemical constituents. However, strong resonances that can be assigned to phenylalanine[23] in the Dromineer spectrum (and to a lesser extent Portumna and Williamstown) are not present in the Coole Bay sample. Phenylalanine is the most commonly found aromatic AA in proteins and enzymes, is invariably present in any animal tissue and is also synthesised by common pathways in phytoplankton and bacteria. It is considered an easily degraded hydrolysable AA,[47] so its presence in some samples is of interest. Phenylalanine has been associated with increased concentrations in water of NH4+,[48] which in turn is a product and indicator of the presence of nitrogenous organic wastes. Dromineer is strongly influenced by the Nenagh River, which passes through land utilised for agriculture and raising livestock, and also accommodates a sizable public marina. Higher phenylalanine concentrations may therefore be an indicator of elevated organic wastes from agriculture and industry. Interestingly, there appears to be little phenylalanine in the Coole Bay sample, which is south of the Dromineer sampling site. This may be explained by the fact that the site is secluded, surrounded by forestry and is not fed or influenced directly by a river. However, during the sampling period from August to September; an exotic invasive species in Ireland, Zebra mussels (Dreissena polymorpha) were evident at highest concentrations on the eastern side of the lake at Coole Bay. The filtering activities of zebra mussels have been shown to have a large ecosystem-level influence on nitrogen cycling[49–51] and organic nitrogen concentrations decrease in water columns in microcosms with live zebra mussels.[52] It is therefore possible that the filtering activities of Zebra mussels result in recycling of larger organic nitrogen compounds such as phenylalanine. The presence of formate in both samples suggests a pathway of organic carbon degradation mainly reported for anoxic marine sediments[53] and indicates that anoxic breakdown by various microorganisms takes place in the lake. Formate and other volatile fatty acids (VFAs) are products of hydrolysis and anaerobic fermentation.[54]
|
A broad background hump from lignin often centred at 6.9–7.1 ppm is present in the aromatic regions of Fig. 6. The presence of lignin-type material is confirmed by the intense methoxy signal seen in the HMQC data (Fig. 3). In addition, the conjugated double bonds are likely the result of the presence of carotenoid structures known to be produced by aquatic species and present in freshwater DOM.[12] The fate of carotenoid structures is not well understood despite an estimated net annual production over ~100 × 106 t from photosynthetic organisms alone.[55,56]
Conclusions
Given the influence of terrestrial organic matter on marine DOM and the similarity in the structures of both, it is challenging to assess the source of DOM and whether it is aquatic or terrestrial in origin. The findings here suggest a strong terrestrial input of recalcitrant material. Land management and human activities are important factors influencing the spatial distribution of DOM within the lacustrine environment. The input of plant material is confirmed by the presence of lignin-type signatures, whereas the influence of microbial biomass from either terrestrial or aquatic sources is highlighted by resonances for peptidoglycan and protein. Soil microbes may also contribute to silicon cycling through stable organo-silicon structures within the cells. The study also confirms the presence of CRAM in DOM from an Irish lake, which suggests that it may be globally ubiquitous.
Acknowledgements
The authors thank the Science Foundation of Ireland (GEOF509), the Irish Environmental Protection Agency (STRIVE program), the Geological Survey of Ireland, the Natural Science and Engineering Research Council of Canada (Discovery Grant, A.J.S.) and the government of Ontario [Early Researcher Award (A.J.S.)] for funding. Thank you also to the anonymous reviewers for the very helpful suggestions and criticisms.
References
[1] J. I. Hedges, R. G. Keil, R. Benner, What happens to terrestrial organic matter in the ocean? Org. Geochem. 1997, 27, 195.| What happens to terrestrial organic matter in the ocean?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXotVektA%3D%3D&md5=46ed4056c34edc962858a6ba71cd0f64CAS |
[2] R. Benner, B. Benitez-Nelson, K. Kaiser, R. M. W. Amon, Export of young terrigenous dissolved organic carbon from rivers to the Arctic Ocean. Geophys. Res. Lett. 2004, 31, L05305.
| Export of young terrigenous dissolved organic carbon from rivers to the Arctic Ocean.Crossref | GoogleScholarGoogle Scholar |
[3] J. I. Hedges, Global biogeochemical cycles – progress and problems. Mar. Chem. 1992, 39, 67.
| Global biogeochemical cycles – progress and problems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38XmsFOmurY%3D&md5=5153737a53530d313d1680ca2a525343CAS |
[4] C. T. Chiou, R. L. Malcolm, T. I. Brinton, D. E. Kile, Water solubility enhancement of some organic pollutants and pesticides by dissolved humic and fulvic-acids. Environ. Sci. Technol. 1986, 20, 502.
| Water solubility enhancement of some organic pollutants and pesticides by dissolved humic and fulvic-acids.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL28XhsFGntb8%3D&md5=82f158ab688b30b53bf394b2f1786f25CAS | 19994935PubMed |
[5] C. D. Keeling, T. P. Whorf, M. Wahlen, J. Van der Plicht, Interannual extremes in the rate of rise of atmospheric carbon dioxide since 1980. Nature 1995, 375, 666.
| Interannual extremes in the rate of rise of atmospheric carbon dioxide since 1980.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXmsFKrtrY%3D&md5=5824bc23dc2ef74c9434411b02621bcbCAS |
[6] T. Dittmar, J. A. Paeng, Heat-induced molecular signature in marine dissolved organic matter. Nat. Geosci. 2009, 2, 175.
| Heat-induced molecular signature in marine dissolved organic matter.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXisVCgt7s%3D&md5=22ddceaae8afdb45988641155abceb25CAS |
[7] R. Stone, The invisible hand behind a vast carbon reservoir. Science 2010, 328, 1476.
| The invisible hand behind a vast carbon reservoir.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXot1yru7c%3D&md5=284dfc5251222deee3262d1a2a248407CAS | 20558685PubMed |
[8] L. A. Cardoza, A. K. Korir, W. H. Otto, C. J. Wurrey, C. K. Larive, Applications of NMR spectroscopy in environmental science. Prog. Nucl. Mag. Res. Sp 2004, 45, 209.
| Applications of NMR spectroscopy in environmental science.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVahsrjK&md5=d07d965034e5166f93c2e2e9751a3967CAS |
[9] A. J. Simpson, Multidimensional solution state NMR of humic substances: a practical guide and review. Soil Sci. 2001, 166, 795.
| Multidimensional solution state NMR of humic substances: a practical guide and review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXovVertr0%3D&md5=f65b1b50171429bdbfaa3a100c52a413CAS |
[10] N. Hertkorn, A. Kettrup, Molecular level structural analysis of natural organic matter and of humic substances by multinuclear and higher dimensional NMR spectroscopy, in Use of Humates to Remediate Polluted Environments: From Theory to Practice (Eds I. V. Perminova, N. Hertkom, K. Hatfield) 2005, pp. 391–435 (Springer: Dordrecht).
[11] B. P. Kelleher, A. J. Simpson, Humic substances in soils: are they really chemically distinct? Environ. Sci. Technol. 2006, 40, 4605.
| Humic substances in soils: are they really chemically distinct?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XmsVaisrg%3D&md5=7252a08f71cae288d32b1caec2821d1bCAS | 16913113PubMed |
[12] B. Lam, A. Baer, M. Alaee, B. Lefebvre, A. Moser, A. Williams, A. J. Simpson, Major structural components in freshwater dissolved organic matter. Environ. Sci. Technol. 2007, 41, 8240.
| Major structural components in freshwater dissolved organic matter.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1OhsLfI&md5=fbd605818c422c362ccd1983734e8d33CAS | 18200846PubMed |
[13] N. Hertkorn, R. Benner, M. Frommberger, P. Schmitt-Kopplin, M. Witt, K. Kaiser, A. Kettrup, J. I. Hedges, Characterization of a major refractory component of marine dissolved organic matter. Geochim. Cosmochim. Acta 2006, 70, 2990.
| Characterization of a major refractory component of marine dissolved organic matter.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XlsFGjurc%3D&md5=3eda8ef3992e1afa104d750b24f73dddCAS |
[14] A. J. Leenheer, Comprehensive approach to preparative isolation and fractionation of dissolved organic carbon from natural waters and wastewaters. Environ. Sci. Technol. 1981, 15, 578.
| Comprehensive approach to preparative isolation and fractionation of dissolved organic carbon from natural waters and wastewaters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3MXlvFaju7Y%3D&md5=98deaa98c038d612dab8585eadd7faf3CAS |
[15] E. M. Thurman, R. L. Malcolm, Preparative isolation of aquatic humic substances. Environ. Sci. Technol. 1981, 15, 463.
| Preparative isolation of aquatic humic substances.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3MXksV2jsL4%3D&md5=bf88220de0f42105a0fe0bcbad714574CAS |
[16] J. P. Simjouw, E. C. Minor, K. Mopper, Isolation and characterization of estuarine dissolved organic matter: comparison of ultrafiltration and C18 solid-phase extraction techniques. Mar. Chem. 2005, 96, 219.
| Isolation and characterization of estuarine dissolved organic matter: comparison of ultrafiltration and C18 solid-phase extraction techniques.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmvFGjt7Y%3D&md5=fbb7c1faa411b4ec426e5df3d601dd8fCAS |
[17] B. Lam, A. J. Simpson, Passive sampler for dissolved organic matter in freshwater environments. Anal. Chem. 2006, 78, 8194.
| Passive sampler for dissolved organic matter in freshwater environments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFSqs7zK&md5=34235a0c8c5a39225834bd6e88bd51e9CAS | 17165807PubMed |
[18] A. J. Simpson, S. A. Brown, N. M. R. Purge, Effective and easy solvent suppression. J. Magn. Reson. 2005, 175, 340.
| Effective and easy solvent suppression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXls1ygsrY%3D&md5=67f12730c50ea4ae54303eca7e7a36cfCAS | 15964227PubMed |
[19] D. Wu, A. Chen, C. S. Johnson, An improved diffusion ordered spectroscopy experiment incorporating bipolar-gradient pulses. J. Magn. Reson. A 1995, 115, 260.
| An improved diffusion ordered spectroscopy experiment incorporating bipolar-gradient pulses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXnsVGht7k%3D&md5=28d633ba0e82d356731b877285874b2fCAS |
[20] S. J. Joseph, P. Hugenholtz, P. Sangwan, C. A. Osborne, P. H. Janssen, Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 2003, 69, 7210.
| Laboratory cultivation of widespread and previously uncultured soil bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXpvFClsrY%3D&md5=216e5f62f1fe32924379c39e3c00c136CAS | 14660368PubMed |
[21] P. H. Janssen, P. S. Yates, B. E. Grinton, P. M. Taylor, M. Sait, Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 2002, 68, 2391.
| Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XjsFGqtLo%3D&md5=e4c64113d5b38727d0c866a448c219aeCAS | 11976113PubMed |
[22] B. P. Kelleher, M. J. Simpson, A. J. Simpson, Assessing the fate and transformation of plant residues in the terrestrial environment using HR-MAS NMR spectroscopy. Geochim. Cosmochim. Acta 2006, 70, 4080.
| Assessing the fate and transformation of plant residues in the terrestrial environment using HR-MAS NMR spectroscopy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XotVejsro%3D&md5=1db0120cd8113a9da1cd623f86b98595CAS |
[23] A. J. Simpson, G. Song, E. Smith, B. Lam, E. H. Novotny, M. H. B. Hayes, Unraveling the structural components of soil humin by use of solution-state Nuclear Magnetic Resonance Spectroscopy. Environ. Sci. Technol. 2007, 41, 876.
| Unraveling the structural components of soil humin by use of solution-state Nuclear Magnetic Resonance Spectroscopy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlWmu7%2FE&md5=d735fc3527a93a777b85974d7346a4cdCAS | 17328197PubMed |
[24] B. G. Pautler, A. J. Simpson, D. J. McNally, S. F. Lamoureux, M. J. Simpson, Arctic permafrost active layer detachments stimulate microbial activity and degradation of soil organic matter. Environ. Sci. Technol. 2010, 44, 4076.
| Arctic permafrost active layer detachments stimulate microbial activity and degradation of soil organic matter.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlvVakt7k%3D&md5=9823a26914c4a18f40139b3e37274564CAS | 20459054PubMed |
[25] A. J. Simpson, W. L. Kingery, P. G. Hatcher, The identification of plant derived structures in humic materials using three dimensional NMR spectroscopy. Environ. Sci. Technol. 2003, 37, 337.
| The identification of plant derived structures in humic materials using three dimensional NMR spectroscopy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XovFGhsbk%3D&md5=b0399cd652dbfb378fc8ee789d38533cCAS | 12564906PubMed |
[26] A. J. Simpson, M. J. Simpson, E. Smith, B. P. Kelleher, Microbially derived inputs to soil organic matter: are current estimates too low? Environ. Sci. Technol. 2007, 41, 8070.
| Microbially derived inputs to soil organic matter: are current estimates too low?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtFGnsLvP&md5=692c4f0fd117acb379bc3e6217b46897CAS | 18186339PubMed |
[27] J. Fuhrman, Dissolved free amino acid cycling in an estuarine outflow plume. Mar. Ecol. Prog. Ser. 1990, 66, 197.
| Dissolved free amino acid cycling in an estuarine outflow plume.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXhslSltL8%3D&md5=7225fc45fb5b464717953ba2fe25b80aCAS |
[28] J. I. Hedges, G. Eglinton, P. G. Hatcher, D. L. Kirchman, C. Arnosti, S. Derenne, R. P. Evershed, I. Kogel-Knabner, J. W. de Leeuw, R. Littke, W. Michaelis, J. Rullkotter, The molecularly uncharacterized component of nonliving organic matter in natural environments. Org. Geochem. 2000, 31, 945.
| The molecularly uncharacterized component of nonliving organic matter in natural environments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXosVGiur4%3D&md5=2c3ae2e9f1355e87277d27602140f590CAS |
[29] E. Tanoue, S. Nishiyama, M. Kamo, A. Tsugita, Bacterial membranes: possible source of a major dissolved protein in seawater. Geochim. Cosmochim. Acta 1995, 59, 2643.
| Bacterial membranes: possible source of a major dissolved protein in seawater.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXms1eltLk%3D&md5=a9144113bbbab986a9b357063b20aa48CAS |
[30] H. Ogawa, Y. Amagai, I. Koike, K. Kaiser, R. Benner, Production of refractory dissolved organic matter by bacteria. Science 2001, 292, 917.
| Production of refractory dissolved organic matter by bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjsVSrt78%3D&md5=8401c5b9a163cb149068c30a24cfd717CAS | 11340202PubMed |
[31] S. K. Park, N. S. Hettiarachchy, L. Were, Degradation behaviour of soy protein-wheat gluten films in simulated soil conditions. J. Agric. Food Chem. 2000, 48, 3027.
| Degradation behaviour of soy protein-wheat gluten films in simulated soil conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXktlerurk%3D&md5=57d99f7fb907cbd9e158c6d34d737c02CAS | 10898660PubMed |
[32] R. A. Herman, J. D. Wolt, W. R. Halliday, Rapid degradation of the Cry1F insecticidal crystal protein in soil. J. Agric. Food Chem. 2002, 50, 7076.
| Rapid degradation of the Cry1F insecticidal crystal protein in soil.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XnvVOjur0%3D&md5=957010ae82d0c21bfc48f1aaa55a7bc6CAS | 12428962PubMed |
[33] A. S. Waksman, K. R. N. Iyer, Contribution to our knowledge of the chemical nature and origin of humus. Soil Sci. 1933, 36, 69.
| Contribution to our knowledge of the chemical nature and origin of humus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaA3sXlslWjtA%3D%3D&md5=219633f04b10dc2f394574b80e8e561aCAS |
[34] K. M. Holtman, H.-M. Chang, H. Jameel, J. F. Kadla, Elucidation of lignin structure through degradative methods: comparison of modified DFRC and thioacidolysis. J. Agric. Food Chem. 2003, 51, 3535.
| Elucidation of lignin structure through degradative methods: comparison of modified DFRC and thioacidolysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjtlemsrc%3D&md5=05d5279247ee30db3e22d2f21b9f611dCAS | 12769520PubMed |
[35] A. P. Deshmukh, A. J. Simpson, P. G. Hatcher, Evidence for cross-linking in tomato cutin using HR-MAS NMR spectroscopy. Phytochemistry 2003, 64, 1163.
| Evidence for cross-linking in tomato cutin using HR-MAS NMR spectroscopy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXotVeqt7w%3D&md5=496bc8e35293781cdfcb1643aac4e9f3CAS | 14568084PubMed |
[36] A. J. Simpson, M. J. Simpson, W. L. Kingery, B. A. Lefebvre, A. Moser, A. J. Williams, M. Kvasha, B. P. Kelleher, The application of 1H high-resolution magic-angle spinning NMR for the study of clay-organic associations in natural and synthetic complexes. Langmuir 2006, 22, 4498.
| The application of 1H high-resolution magic-angle spinning NMR for the study of clay-organic associations in natural and synthetic complexes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjtFWrtLo%3D&md5=df2535ff0fcb2683bb0d66c409b697e8CAS | 16649755PubMed |
[37] H. W. Zhang, D. W. Niesel, J. W. Peterson, G. R. Klimpel, Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 1998, 66, 5196..
| 9784522PubMed |
[38] P. Tréguer, D. M. Nelson, A. J. van Bennekorn, D. J. DeMaster, A. Leynaert, B. Quéguiner, The silica balance in the world ocean: a re-estimate. Science 1995, 268, 375.
| The silica balance in the world ocean: a re-estimate.Crossref | GoogleScholarGoogle Scholar | 17746543PubMed |
[39] O. Ragueneau, P. Treguer, A. Leynaert, R. F. Anderson, M. A. Brzezinski, D. J. De Master, R. C. Dugdale, J. Dymond, G. Fische, R. Francois, C. Heinze, E. Maier-Reimer, V. Martin- Jezequel, D. M. Nelson, B. Queguiner, A review of the Si cycle in the modern ocean: recent progress and missing gaps in the application of biogenic opal as paleoproductivity proxy. Global Planet. Change 2000, 26, 317.
| A review of the Si cycle in the modern ocean: recent progress and missing gaps in the application of biogenic opal as paleoproductivity proxy.Crossref | GoogleScholarGoogle Scholar |
[40] A. Yool, T. Tyrrell, Role of diatoms in regulating the ocean’s silicon cycle. Global Biogeochem. Cycles 2003, 17, 1103.
| Role of diatoms in regulating the ocean’s silicon cycle.Crossref | GoogleScholarGoogle Scholar |
[41] D. J. Conley, Terrestrial ecosystems and the global biogeochemical silica cycle. Global Biogeochem. Cycles 2002, 16, 1121.
| Terrestrial ecosystems and the global biogeochemical silica cycle.Crossref | GoogleScholarGoogle Scholar |
[42] P. W. Birkeland, Soils and Geomorphology 1999, 3rd edn (Oxford University Press: New York).
[43] N. van Breemen, P. Buurman, Soil Formation 2002 (Kluwer Academic Press: Dordrecht).
[44] M. Sommer, D. Kaczorek, Y. Kuzyakov, J. Breuer, Silicon pools and fluxes in soils and landscapes: a review. J. Plant Nutr. Soil Sci. 2006, 169, 310.
| Silicon pools and fluxes in soils and landscapes: a review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xmtlymsro%3D&md5=767702ab3b25e9cb65fe19fd56969d84CAS |
[45] R. Brindle, M. Punch, K. Albert, H MAS NMR spectroscopy of chemically modified silica gels: a fast method to characterize stationary interphases for chromatography. Solid State Nucl. Mag. 1996, 6, 251.
| H MAS NMR spectroscopy of chemically modified silica gels: a fast method to characterize stationary interphases for chromatography.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XktlShuro%3D&md5=7e96e2a9bed100e4ed1e61213fc116f7CAS |
[46] B. Lam, A. J. Simpson, Direct 1H NMR spectroscopy of dissolved organic matter in natural waters. Analyst (Lond.) 2008, 133, 263.
| Direct 1H NMR spectroscopy of dissolved organic matter in natural waters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVGnur0%3D&md5=f6197adfbecc340596338d571ffc3a26CAS |
[47] Y. Yamashita, E. Tanoue, Distribution and alteration of amino acids in bulk DOM along a transect from bay to oceanic waters. Mar. Chem. 2003, 82, 145.
| Distribution and alteration of amino acids in bulk DOM along a transect from bay to oceanic waters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXmtFamtbc%3D&md5=2586fd4d24002da404c1f2f827a0fc72CAS |
[48] A. B. Jones, W. C. Dennison, G. R. Stmart, Macroalgal responses to nitrogen source and availability: amino acid metabolic profiling as a bioindicator using Gracilaria edulis (Rhodophyta). J. Phycol. 1996, 32, 757.
| Macroalgal responses to nitrogen source and availability: amino acid metabolic profiling as a bioindicator using Gracilaria edulis (Rhodophyta).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXitFGitg%3D%3D&md5=a61b65980d974a294df03d8a5e7c1cedCAS |
[49] E. Mellina, J. B. Rasmussen, E. L. Mills, Impact of zebra mussel (Dreissena polymorpha) on phosphorus cycling and chlorophyll in lakes. Can. J. Fish. Aquat. Sci. 1995, 52, 2553.
| Impact of zebra mussel (Dreissena polymorpha) on phosphorus cycling and chlorophyll in lakes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xitl2gsrs%3D&md5=cf1e7d2cde7c3f57d0ec3462ac985484CAS |
[50] D. L. Arnott, M. J. Vanni, Nitrogen and phosphorus recycling by the zebra mussel (Dreissena polymorpha) in the western basin of Lake Erie. Can. J. Fish. Aquat. Sci. 1996, 53, 646.
| Nitrogen and phosphorus recycling by the zebra mussel (Dreissena polymorpha) in the western basin of Lake Erie.Crossref | GoogleScholarGoogle Scholar |
[51] J. D. Conroy, W. J. Edwards, R. A. Pontius, D. D. Kane, H. Zhang, J. F. Shea, J. N. Richey, D. A. Culver, Soluble nitrogen and phosphorus excretion of exotic freshwater mussels (Dreissena spp.): potential impacts for nutrient remineralisation in western Lake Erie. Freshw. Biol. 2005, 50, 1146.
| Soluble nitrogen and phosphorus excretion of exotic freshwater mussels (Dreissena spp.): potential impacts for nutrient remineralisation in western Lake Erie.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXms1aqsL8%3D&md5=f99511e421eb9f6cb35f893c1c1fb57eCAS |
[52] O. Bykova, A. Laursen, V. Bostan, J. Bautista, L. McCarthy, Do zebra mussels (Dreissena polymorpha) alter lake water chemistry in a way that favours Microcystis growth? Sci. Total Environ. 2006, 371, 362.
| Do zebra mussels (Dreissena polymorpha) alter lake water chemistry in a way that favours Microcystis growth?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFOrtbjF&md5=67228a5f7e665aebd0debc41a36e873eCAS | 17011023PubMed |
[53] D. J. Burdige, Sediment pore waters, in Biogeochemistry of Marine Dissolved Organic Matter (Eds D. A. Hansell, C. A. Carlson) 2002, vol. 13, p. 637 (Elsevier Science: New York).
[54] K. Mopper, D. J. Kieber, Distribution and biological turnover of dissolved organic compounds in the water column of the Black Sea. Deep-Sea Res. 1991, 38, S1021.
| Distribution and biological turnover of dissolved organic compounds in the water column of the Black Sea.Crossref | GoogleScholarGoogle Scholar |
[55] J. W. Louda, L. Liu, E. W. Baker, Senescence- and death-related alteration of chlorophylls and carotenoids in marine phytoplankton. Org. Geochem. 1635, 2002, 12..
[56] T. Matsuno, New structures of carotenoids in marine animals. Pure Appl. Chem. 1985, 57, 659.
| New structures of carotenoids in marine animals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2MXktlGjsLc%3D&md5=c0db95a20e34eb93d0db3f4b03cb5304CAS |


