Assessing plant translocation success: common metrics mask high levels of inbreeding in a recently established Banksia brownii (Proteaceae) population
Rebecca Dillon A B * , David Coates A C , Rachel Standish D , Leonie Monks
A B * , David Coates A C , Rachel Standish D , Leonie Monks  A D and Michelle Waycott B
A D and Michelle Waycott B
A Biodiversity and Conservation Science, Department of Biodiversity, Conservation and Attractions, Locked Bag 104, Bentley Delivery Centre, Bentley, WA 6983, Australia.
B School of Biological Sciences, The University of Adelaide, Adelaide, SA 5005, Australia.
C School of Biology, University of Western Australia, 35 Stirling Highway, Perth, WA 6009, Australia.
D Environmental and Conservation Sciences, Murdoch University, 90 South Street, Murdoch, WA 6150, Australia.
Australian Journal of Botany 71(2) 79-92 https://doi.org/10.1071/BT22071
Submitted: 5 July 2022 Accepted: 7 February 2023 Published: 17 March 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: As threatening processes continue to impact rare plant populations, the use of translocations is becoming increasingly frequent. The ultimate success of translocation, attaining long-term persistence, is determined by species’ ability to reproduce, recruit, and maintain levels of genetic diversity that permits the capacity to adapt to environmental change.
Aims: This study aimed to assess translocation success of Banksia brownii.
Methods: We compared genetic diversity, reproductive output and aspects of the mating system of a translocated population with four reference wild populations.
Key results: We found that the use of multiple source populations for the translocation resulted in levels of genetic diversity comparable to reference populations of high diversity. Reproductive output was highest in the translocated population and a positive relationship between plant size and number of flowers across all populations was evident. However, mating system analysis revealed a large difference in outcrossing rates between populations in different habitats despite common pollinators, with the translocated population having the highest rates of selfing.
Conclusion: Whilst genetic diversity and reproductive output were comparable or higher in the translocated population than in the reference populations, assessing translocation outcomes by these measures alone can potentially mask the longer-term erosion of genetic diversity through unfavourable patterns of mating.
Implications: We recommend using multiple metrics of reproductive, recruitment and genetic diversity to properly assess plant translocation success.
Keywords: Banksia, genetic diversity, mating systems, pollination, reintroduction, reproductive output, threatened plant, translocation, Western Australia.
Introduction
The use of translocations, as defined by Commander et al. (2018) and IUCN/SSC (2013), in plant species conservation is becoming increasingly frequent as threats such as land clearing, habitat fragmentation, rapid environmental change, diseases, and invasive species intensify and become progressively more difficult to manage (Godefroid et al. 2011). Despite the increasing focus and the numerous attempts worldwide, there are still relatively few examples demonstrating plant translocation success through the establishment of a viable population and long-term (beyond 10 years) population persistence (Seddon et al. 2007; Wendelberger et al. 2008; Godefroid et al. 2011; Lawrence and Kaye 2011; Guerrant 2013; Commander et al. 2018; Silcock et al. 2019).
While the ultimate goal of a translocation is to achieve long-term persistence of a population without the need for further human intervention (Seddon 1999), initial goals often include plant establishment evaluated through the assessment of plant survival and growth. Beyond this, the long-term persistence of translocated populations is determined by their ability to reproduce, recruit, and maintain adequate genetic diversity that permits resilience and the capacity to adapt to change such as declining rainfall or altered disturbance regime (Menges 2008).
Genetic diversity is a key consideration for the success of plant translocations (Weeks et al. 2011). Translocated populations are susceptible to adverse genetic issues such as founder effects, genetic drift and inbreeding, largely due to their small size, but also as a result of commonly being sourced from wild populations that may have a history of fragmentation and reductions or bottlenecks in population size (Jachowski et al. 2016). Several meta-analyses have demonstrated a positive relationship between genetic diversity, population size and fitness (e.g. Reed and Frankham 2003; Leimu et al. 2006) emphasising that loss of genetic diversity and inbreeding are particularly pronounced in small populations. Translocated populations established from limited genetic material may experience a reduction in fitness associated with inbreeding depression. In plants this is often expressed as reduced germination, survival and reproductive output (Llorens et al. 2013). In the long term, genetic diversity enables adaptive evolutionary change contributing to species persistence (Guerrant and Kaye 2007; Keller et al. 2012).
The establishment of new populations of threatened species is often compromised by using propagules from genetically impoverished source populations (Frankham 2015; Weeks et al. 2016; Liddell et al. 2020). To overcome this limitation, Ralls et al. (2018) emphasise that the rationale for the mixing of gene pools follows the general principles of ‘genetic rescue’. They argue that the negative effects of inbreeding and low genetic diversity can often be ameliorated or nullified by combining gene pools representing genetically different populations. To date, several studies have shown translocated populations obtain levels of diversity equivalent to or higher than their source, largely attributed to mixing source populations and the number (and genetic diversity) of the founders used (Ramp et al. 2006; Lloyd et al. 2012; Fant et al. 2013; Alonso et al. 2014; Zavodna et al. 2015). However, studies linking such results with tangible increases in fitness in translocated populations, particularly in those species that are long lived, are scarce (but see Van Rossum et al. 2020).
Whilst maximising genetic diversity of founder populations is considered key to translocation success, maintaining that genetic diversity over future generations is equally as important, with the genetic quality of progeny promoting population persistence (Aguilar et al. 2019). Genetic loss can occur at all stages of translocation (Fant et al. 2013; St. Clair et al. 2020). Following the initial establishment of founders, the genetic diversity of their progeny and the maintenance of genetic diversity will be significantly influenced by several factors. These include founding population size; the survival of individuals to reproductive age; reproductive output; functioning of the mating system and associated factors including pollinator availability and behaviour at the translocation site (Monks et al. 2012; Whitehead et al. 2018).
Mating systems, in particular, can be useful indicators of population processes and can give valuable insight into the development of management strategies for conservation (Coates et al. 2007; Eckert et al. 2009) and, translocation and restoration more specifically (Monks et al. 2012; Ritchie and Krauss 2012; Millar et al. 2019a, 2019b). Plants exhibit a wide array of mating systems, from predominant selfing to predominant outcrossing, with contrasting effects on patterns of genetic variation and rates of inbreeding within individual species (Brown 1979; Hamrick and Godt 1989). Plant mating systems can be affected by self-incompatibility systems and the attributes of the individual plant such as size, floral structure and display, and also by ecological variables such as pollination (e.g. pollinator assembly, abundance and foraging behaviour), population size, sex ratios (in non-hermaphroditic species), spatial structure, and density and position in the landscape (Coates et al. 2007; Sampson et al. 2016). Small and/or isolated populations are predicted to have decreased levels of outcrossing and increased levels of mating between relatives (Aguilar et al. 2006). This is likely to lead to loss of genetic diversity through drift and for predominately outcrossing species, the likely expression of inbreeding depression (Dudash et al. 2005; Ottewell et al. 2016).
Pollinator availability and behaviour may be altered in translocated populations due to their small size (Oostermeijer et al. 2000) or because of establishment of a new population in habitat where pollinator assemblages differ or are absent. As a result, the effect translocation may have on plant mating system function is unknown but changes due to pollinator availability and behaviour in animal pollinated species, population size and spatial structure are likely to be particularly relevant. The patterns of pollination are likely to affect the maintenance of genetic diversity through aspects of the mating system by determining the levels of outcrossing or inbreeding and the degree to which plants receive pollen from the same plant (Ghazoul 2005; Llorens et al. 2013).
Recent assessments of the success of restored populations of common species have included comparisons of genetic diversity, mating system parameters and reproductive output between wild and restored sites and inferences on pollinator presence and behaviour (Ritchie and Krauss 2012; Millar et al. 2019a, 2019b, 2020). Whilst studies on threatened plant translocations investigate some of these factors, very few examine all three factors together. The study of genetic diversity levels of translocated populations, the mating systems operating within them, and any effect on reproductive output can contribute to the understanding of factors crucial to population persistence, and importantly contribute to a prediction of a populations’ persistence into the future (Millar et al. 2019a).
Over the past three decades proteaceous species with a range of animal pollinators have been established at over 30 translocation sites in south-western Western Australia (DBCA, unpubl. data). Several of these projects have achieved the goals of survival, growth and reproduction at rates comparable to wild populations. These populations provide an opportunity for further evaluation of translocation success through the assessment of genetic diversity capture, maintenance of mating system function and reproductive output. Banksia brownii Baxter ex R. Br. is a threatened proteaceous shrub that has been targeted for translocation in Western Australia and that provides a model system for investigation into the effects of translocation on genetic diversity, mating systems and reproductive output. The species occurs in both upland and lowland habitats and significant genetic structure has been revealed between these habitats (Coates et al. 2015). Widespread decline in both upland and lowland groups has been documented, largely due to the effects of Phytophthora dieback disease and inappropriate fire intervals. The augmentation of existing populations is not feasible due to these threats and as such introductions both within and outside the species range have been undertaken with varied success. A small lowland translocation established in 2007, has been deemed particularly successful using standard metrics to assess short to medium-term success, with high survival and growth rates and the production of fruit annually from 2 years of age.
We investigated genetic diversity, mating systems and reproductive output of a translocated population of B. brownii and compared these measures to those of both large and small wild reference populations across the species range. Additionally, the relative contribution to seed set by insect and vertebrate pollinators at each site was assessed to aid in the interpretation of any differences found in mating system or reproductive output results. Specifically, we address the following questions: (1) how do levels of genetic diversity in a mixed source translocated population compare to the those of the individual source populations and also to both large (>15 000 plants) and small (<200 plants) populations across the species range?; (2) how do mating system parameters in the translocated population compare to those in wild reference populations?; (3) do contributions of invertebrate and vertebrate pollinators differ in response to population size or habitat?; and (4) how do measures of reproductive output in the translocated population compare to those in wild populations? Additionally, the study also provides an opportunity to investigate temporal changes in genetic diversity levels in translocated and wild populations.
Materials and methods
Study species
B. brownii is endemic to south-west Western Australia where it exists in three geographically disjunct population clusters over a linear range of approximately 90 km. Much of the lowland area in which the species occurs has been cleared for agriculture and populations are restricted to vegetation remnants (Sampson et al. 1994). Contemporary decline in population size is attributed to Phytophthora dieback disease and inappropriate fire regimes (Gilfillan and Barrett 2005). It was originally recorded from 30 populations within this range but ten of these populations have been extinct since 1996 due to Phytophthora dieback. Of the extant populations, 13 are close to extinction, with fewer than five mature plants remaining in each. The species is ranked as Endangered under the Australian Government’s Environment Protection and Biodiversity Conservation Act 1999 and Critically Endangered under the Western Australian Biodiversity Conservation Act 2016 (Gilfillan and Barrett 2005).
B. brownii occurs in floristically-rich heathy shrublands and woodlands dominated by species from the Proteaceae, Myrtaceae and Fabaceae. It is a long-lived small tree or shrub usually growing to 2–3 m in height. In some elevated areas of the Stirling Range (500–1100 m altitude) it grows as a low shrub to 1.5 m and in more sheltered sites as an openly branched small tree to 6 m (Rye and Hopper 1981). Its cylindrical inflorescences (Supplementary Fig. S1) contain an average of 1249 protandrous flowers (Collins et al. 1993) that are produced between March and July (Day et al. 1997). Honeyeaters, honey possums, rodents and insects have been observed foraging on the flowers (Day et al. 1997). The species is self-compatible and exhibits a mixed mating system with substantial levels of self-pollination (Sampson et al. 1994). Pollinated inflorescences develop into infructescences; woody ‘cone’ structures that are embedded with follicles. Each follicle usually contains one black, triangular seed with a papery wing. Follicle opening and seed release generally occurs in response to fire.
Significant genetic structure has been revealed within B. brownii using microsatellite genotyping, which indicates three genetically distinct units (Coates et al. 2015). These units are geographically separated: an upland unit confined to the Stirling Range (six extant populations), a central lowland unit between Albany and the Stirling Range (five extant populations) and a southern lowland unit on the Vancouver Peninsula (represented by a single population) south of Albany. The genetic and ecological distinction among units has resulted in each requiring separate management in terms of translocation (Coates et al. 2015).
Recovery actions for the species to date have included the establishment of three translocated populations (one for each of the recognised conservation units), at sites free from threatening processes such as Phytophthora dieback. The translocated population representing the central lowland unit was established in 2007/8 and has been the most successful in terms of survival and reproductive output. It forms the basis of this study. The remaining translocated populations could not be included due to population age (not reproductive at time of sampling) and the effect of drought on seedling survival (insufficient number of plants to sample).
Translocated population
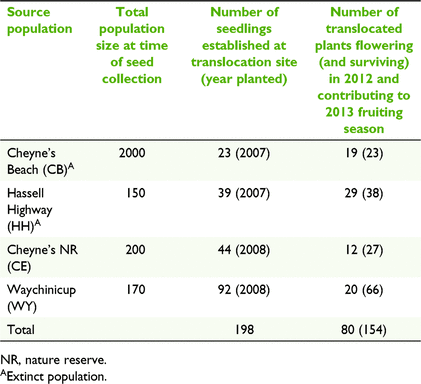
The translocation site is located within a 175-ha patch of remnant vegetation similar to that found at the source populations. It is approximately 10 km to the north-east of the nearest source population. Seedlings were grown from seed and maintained under nursery conditions until approximately 6 months of age. In the winter of 2007, 62 seedlings from the two extinct source populations were established and in 2008, an additional 136 seedlings from the two extant source populations were planted at the site (Table 1). All seedlings were genotyped prior to planting using microsatellite markers as part of study investigating genetic structure and genetic diversity loss following population extinction (Coates et al. 2015). Seedlings were planted so that populations were mixed, and siblings were separated spatially. In the 5 years following planting, survival and growth were high. Plants began to flower in 2010, at 2–3 years of age and in the year prior to population sampling (2012), 52% of the 154 surviving founders were flowering. These individuals are considered to have contributed to the 2013 fruiting season (Table 1).
|
Source populations
The translocated population was established from seedlings grown from seeds sourced from four populations within the central lowland unit. The four central lowland source populations were located within a range of 12 km of the translocation site. Two of these populations have been extinct for over 20 years, thus seedlings for translocation were grown from seed collected in 1989 and stored at the Western Australian Seed Centre, Kensington at −18°C and 15% relative humidity. At the time of seed collection all populations were in decline and three of the four contained 200 or less plants (Table 1).
Genotype data for the four source populations was generated as part of a previous study (Coates et al. 2015) and combined and re-analysed here to estimate the genetic diversity of the founder population.
Wild populations
Comparing levels of genetic diversity, mating system parameters and reproductive output of this translocated population with those of wild populations can allow us to predict whether genetic diversity is likely to be maintained and thus the likelihood of success over the longer term. As the translocated population is small, levels of genetic diversity and patterns of pollination that are comparable to small reference populations may be expected, although alignment with those of large healthy populations may be more desirable and indicate the capture of adequate diversity and acquisition of functional processes within the translocated population. Coates et al. (2015) found that the total genetic diversity in the species to be highest in the upland unit with up to 75% of the total estimated genetic diversity in B. brownii found in the Stirling Range populations. We have therefore included two upland populations in our study to give an indication of how the translocated population is performing in comparison not only to central lowland populations, but also to populations of known high genetic diversity. Including populations from across the species range also allows us to investigate the occurrence of any differences in mating system function between upland and lowland groups.
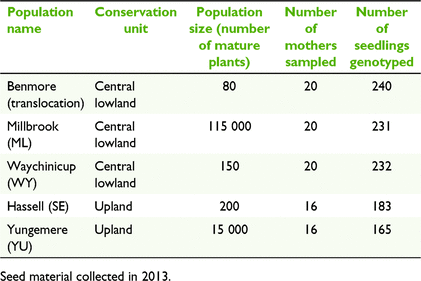
For comparison of population performance with the translocated population, four wild populations were selected for this study representing both a large and small population from each of the upland unit and the central lowland unit (Fig. 1; Table 2).
|
|
|
Population sampling
Four to six mature cones were collected from 16 to 20 individuals from each of the five populations (Table 2). Cones were a result of the 2013 fruiting season. In wild populations, to minimise collection from related individuals, sampling targeted plants spread throughout the population and avoided nearest neighbours. In the translocated population, all fruiting plants were sampled. Cones from each individual tree were kept separate.
Seed was extracted from cones by burning the cones with a portable gas torch, soaking in water for 24 h and drying on trays. Fifteen seed from each maternal tree were randomly selected and plated on agar, individually labelled, and germinated at 15°C. When cotyledons had emerged, seedlings were transferred to potting mix in trays and grown on under nursery conditions. At 4 months of age, leaf samples from 10 to 15 seedlings per mother were collected, freeze dried and stored on silica. A representative specimen was also collected from each of the five sampled populations, identification was verified, and the specimens were lodged at the Western Australian Herbarium (PERTH 03124029, PERTH 07756968, PERTH 08209154, PERTH 03127877, PERTH 08397244). Genomic DNA was extracted from leaf material following the methods of McArthur and Coates (2010).
Utility of loci
A total 1051 seedlings were genotyped for genetic diversity and mating systems analysis using eight nuclear microsatellite markers (loci A1027, A11, B1027, B1148, A1072, D86, B5 and B7) described previously Coates et al. (2015). These loci are known to have a low frequency of null alleles, numbers of alleles ranging from 7 to 20 per locus, and no locus pairs in linkage disequilibrium across any population. The eight highly polymorphic loci were sufficient for detailed mating system analysis (Ritland 2002), and for assessing patterns of genetic variation within and among populations (Coates et al. 2015).
Alleles were sized using GENEMAPPER v4.0 (Applied Biosystems, Foster City, CA, USA) and double-checked manually. When necessary, the alleles were manually adjusted. Samples that did not amplify well were re-amplified at least once.
Genetic diversity
Genetic diversity parameters effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He) and the Fixation index (Fis) were estimated using GENALEX v6.5 (Peakall and Smouse 2012) and allelic richness (Nar) was calculated using HP-RARE (Kalinowski 2005) based on genotypes for the eight loci for the founding plants at Benmore and adult plants from the same wild populations as this study, sampled in 2006, and also based on genotypes for the eight loci for the 2013 progeny from each population. Comparisons in the levels of genetic diversity parameters between the translocated population and its source populations, between translocated and large and small, and between upland and lowland populations groups at the population level, were assessed using a one-way analysis of variance (ANOVA) with values for each locus used as replicates, conducted in Microsoft Excel software. Where a significant (P < 0.05) difference was observed, means were separated using Tukey HSD post hoc tests.
Multilocus pairwise differentiation of populations based on 2013 progeny was estimated using unbiased FST and DEST in GENALEX 6.5 (Peakall and Smouse 2012) based on the G-statistic option with significance tested using 999 permutations.
Mating system
Mating system parameters for each population were estimated using all eight loci and analysed under both the mixed and correlated mating models using MLTR 3.4 (Ritland and Jain 1981; Ritland 2002). The expectation-maximisation (EM) method was used to estimate the means and standard deviations of the multi-locus outcrossing rate (tm) and single-locus outcrossing rate (ts), estimated rate of bi-parental inbreeding (tm − ts), correlation of selfing among maternal plants (rs) and multi-locus correlated paternity (rp) (Ritland 2002). The standard deviation of each estimate was based on 1000 bootstraps. The effective number of pollen donors in each population was estimated as NeP = 1/rp (Ritland 1989). Family estimates of ts and tm were used to assess differences between translocated and large and small populations, and between upland and lowland populations groups using a one-way ANOVA and Tukey’s HSD post hoc tests in Microsoft Excel. Data with values constrained between 0 and 1 were log transformed prior to analysis. Pairwise z-tests calculated in Microsoft Excel were used to assess differences between bi-parental inbreeding (tm − ts) and correlated paternity (rp) for each population pair.
Pollinator exclusion study
Fifteen reproductive trees, each bearing a minimum of five inflorescences, were randomly selected at four populations (Hassell, Millbrook, Waychinicup and the translocated population) for the pollinator exclusion study. The fifth population, Yungemere, was excluded as flowers were already opening at the time of visitation. On each tree, three inflorescences still in bud, of similar size and located at similar heights in the canopy, were selected and the branch supporting them labelled. Inflorescences were randomly assigned as exposed controls or covered with bags or cages. Cages were designed to permit invertebrate pollinator access but exclude vertebrates and were constructed of aviary mesh with apertures of 10 mm. Cages were large enough to provide adequate space around the fully opened inflorescence to prevent vertebrates attempting to access nectar through the mesh. Breathable, fine mesh (12 threads per cm) nylon bags were used to exclude relevant animal pollinators. These were either dark brown or green in colour to minimise impact on pollinator behaviour. Bags were placed over the inflorescence and arranged in a manner to minimise contact with the plant and tied at the base with a draw string. Cones from the labelled branches were collected 12 months later, and the number of follicles recorded for each.
Reproductive output
Reproductive output was measured at each of the five populations with 20 reproductive trees randomly selected and tagged. The number of inflorescences were counted in year 1 (2017) and the number of cones counted in year 2 (2018) on each tree. The mean percentage of inflorescences that developed into cones were calculated for each population. Three mature cones were collected from each tree. Cone length and the number of follicles on each cone were recorded. These data were used to calculate the mean number of follicles per cone and the mean number of follicles per centimetre of cone, to account for differences in cone length between populations. The number of follicles is a reliable predictor of seed production because each follicle has one seed on average. This was confirmed from a subset of five cones from each population with the estimate of mean seed per follicle found to be 0.97–0.99 across all five sites. Thus, an estimate of the mean number of seed per plant for each population was calculated as the mean number of cones per plant × mean number of follicles per plant.
Canopy heights (from ground to apex in m) and maximum widths (widest width (w1) and width perpendicular to w1 (w2) in m) were measured for each tree and their volumes calculated using the equation for the volume of a cone = ((1/3) × π × [w1 × 0.5]2 × height). These volumes were then used to assess any effect of canopy size on the number of flowers produced.
Analyses of variance (ANOVA) were used to assess variation between pollinator treatments, in canopy volume, the number of flowers per plant, cone length and mean number of follicles per cone between populations and also between upland and lowland, and large and small population groups. Where a significant (P < 0.05) difference was observed, means were separated using Tukey’s HSD post hoc tests. The correlation between the mean number of inflorescences produced and mean canopy size, and also multi-locus outcrossing rate and mean number of inflorescences, was assessed by using Pearson’s correlation test calculated in SOCSCISTATISTICS (https://www.socscistatistics.com/tests/pearson/default2.aspx).
Results
Genetic diversity
Translocated population and its source populations
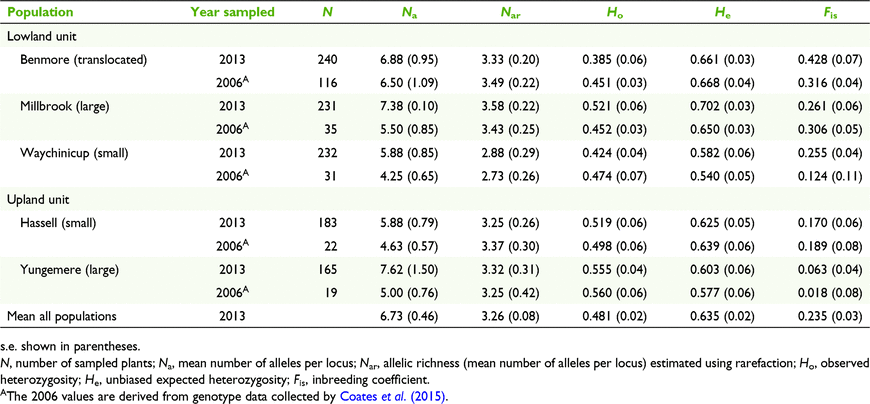
The number of alleles per locus ranged from 3 to 12 (mean = 6.5) and a total of 52 alleles were detected across eight loci for 116 individuals from the four source populations. Combining the genotype data of the four source populations to represent the translocated population resulted in the translocated population containing the highest allelic richness (Nar) but not significantly so. Similarly, between the source populations and between translocated and source populations, there were no significant differences in Ne, Ho, and He (P > 0.05). The inbreeding coefficient (Fis) was positive and significantly greater than 0 in all populations. Fis was high in both the translocated population (Fis = 0.316) and CE population (Fis = 0.361) and significantly greater than in the WY and CB populations (P < 0.05). Expected heterozygosity was highest in the HH population (He = 0.670), followed closely by the translocated population (He = 0.668) (Table S1).
Progeny of translocated and wild populations
Overall levels of mean diversity of progeny from seed collected in 2013 from the five populations (Nar 3.26, Ne 3.07, Ho 0.481, He 0.635) were not significantly different to the levels obtained for adults from these populations in 2006 (Table 3). The number of alleles per locus ranged from 7 to 20 (mean = 11.75) and a total of 94 alleles were detected across eight loci for 1051 individuals from five populations. Allelic richness of the translocated population is comparable to that of the large and upland populations. Comparisons between populations, between small and large population groups, and between central lowland and upland population groups, showed no significant differences in Nar, Ne, Ho, and He. The inbreeding coefficient (Fis) was high in the translocated population with a mean Fis of 0.428 and was found to be significantly higher than wild populations (F(4,35) = 5.68, P < 0.01). Within all wild populations, the inbreeding coefficient was positive (ranging between 0.063 and 0.261), with Yungemere exhibiting the lowest value and Millbrook the highest. The Fis values for the upland and central lowland groups also differed significantly (F(1,38) = 13.76, P < 0.01) with values for lowland populations higher than upland. A small, but insignificant rise in Fis values was observed between generations in the translocated and small lowland population.
|
Pairwise estimates of differentiation were moderate among the upland populations (FST 0.081, DEST 0.278) and low among the central lowland populations (FST 0.028–0.051, DEST 0.119–0.188) (Table S2). Pairwise estimates between the upland populations and the central lowland populations (including the translocated population) were high (FST 0.116–0.125, DEST 0.378–0.563). Estimates of pairwise population differentiation were significant at P < 0.001 for all population pairs tested (Table S2).
Mating systems
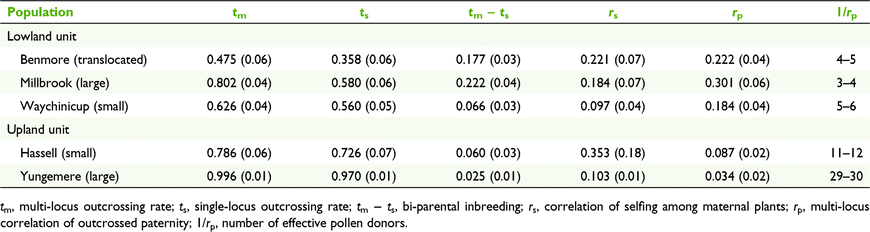
The outcrossing rates varied considerably among the populations and were markedly lower at the translocated population that at any of the reference (wild) populations (Table 4). Outcrossing rates of the five populations ranged from high in the Yungemere population (tm = 0.929, ts = 0.970) to low in the translocated population (tm = 0.475, ts = 0.358) indicating a high degree of selfing in the latter. Based on the ANOVA of family estimates for ts and tm, outcrossing rates were found to differ significantly between populations (F(4,92) = 6.06, P < 0.001). Post hoc Tukey tests revealed that seed families at Yungemere had a significantly higher outcrossing rate compared to all populations (ts = 0.961, P < 0.01, tm = 0.951, P < 0.05) and the translocated population significantly lower compared to all populations (ts = 0.406, P < 0.01, tm = 0.633, P < 0.05), with the exception of the small lowland Waychinicup population. Family estimates of outcrossing rates (tm and ts) were significantly lower in lowland (ts = 0.506, tm = 0.736, compared to upland (ts = 0.903, tm = 0.827) populations (F(1, 90) = 11.41, P < 0.01) and higher in large (ts = 0.725, tm = 0.771) vs small (ts = 0.634, tm = 0.517) populations (F(1, 90) = 11.77, P < 0.01). These significant differences were maintained when the translocated population was removed from the analysis; for population size (F(1, 70) = 5.22, P < 0.05) and for upland vs lowland (F(1,70) = 5.54, P < 0.05). Biparental inbreeding was evident within all populations with the highest levels detected in the Millbrook population (tm − ts = 0.222) and the translocated population (tm − ts = 0.177). Estimates of correlated paternity differed significantly in pairwise comparisons between all upland and lowland populations (z = 2.43–4.49, P < 0.01), with maternal plants in the Stirling Range populations having the greatest number of effective pollen donors (Yungemere Ne = 29–30) and the lowland populations the least (Millbrook Ne = 3–4), while the value for the translocated population (Ne = 4–5) was between the small and large lowland populations.
|
Reproductive output
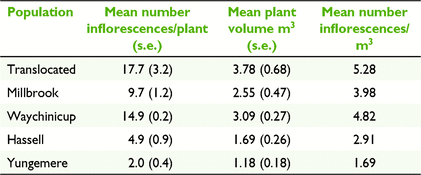
Canopy volume was positively correlated with the number of inflorescences per plant (Pearson’s R = 0.901, d.f. = 3, P < 0.05). Plants in the lowland populations had significantly larger canopy volumes (F(1,107) = 23.87, P < 0.001) and more inflorescences per plant (F(1,107) = 31.13, P < 0.001) compared to the upland populations. Differences in canopy volume and number of inflorescences between large and small population groups were not significant. Plants in the translocated population were on average the largest (3.78 m3) and supported the highest number of inflorescences per unit of canopy volume (5.28 inflorescences/m3). Yungemere had the smallest plants (1.18 m3), which on average held 1.69 inflorescence per m3 of canopy volume (Table 5). Across populations, there was a significantly negative correlation between the mean number of inflorescences per plant and outcrossing rate (Pearson’s R = −0.947, d.f. = 3, P < 0.05) (Fig. 2).
|
|
|
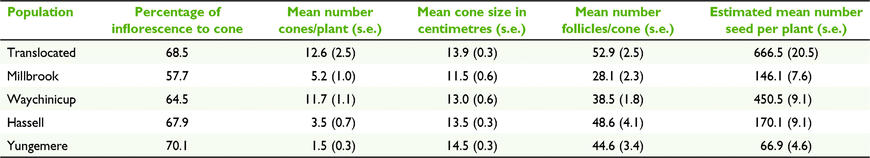
The percentage of inflorescences that produced cones varied among populations but did not differ significantly. Yungemere had the highest conversion rate (70%) and Millbrook the lowest (58%). The mean number of cones per plant was significantly greater (F(4,104) = 12.98, P < 0.001) for the translocated (12.5) and small lowland population (11.7) compared to the remaining populations (1.5–5.2).
The mean length of cones ranged from 11.5 cm (Millbrook) to 14.5 cm (Yungemere) with no significant differences between populations, or large and small, or lowland and upland groups. The mean number of follicles per cone varied from 28.4 (Millbrook) to 52.9 (translocated), with the translocated population differing significantly from the two lowland populations (F(4,196) = 12.20, P < 0.001). Differences in follicle production between large and small, upland and lowland population groups and translocated and other populations were not significant. Accounting for the mean number of cones per plant and the mean number of follicles per cone, the translocated population had a greater estimated mean number of seeds per plant (657.6), significantly different to all populations except Waychinicup (F(4,97) = 12.34, P < 0.001). Yungemere had the lowest output (79.3 seeds per plant) (Table 6).
Calculating the mean number of follicles per centimetre of cone length allowed a standardised assessment of follicle production accounting for differences in cone length, however relative relationships between the values for the populations remained consistent with those obtained for mean number of follicles/cone, as such values are not reported.
Pollinator exclusion study
Across all four populations, 100% of unbagged inflorescences that were open to all animal pollinators produced follicles. In comparison only 10–30% of caged inflorescences, where vertebrates were excluded, produced follicles. Inflorescences that produced follicles where all pollinators were excluded were only found in the Mt Hassell population and constituted 10% (two inflorescences) of those bagged.
The numbers of follicles produced per cone in the control (unbagged) were significantly different to the cage and bagged treatments (F(2,237) = 3.19, P < 0.001) with the mean number of follicles per cone ranging from 28 (Millbrook) to 54 (translocated) for the control and 0.9 (Millbrook) to 3.4 (Mt Hassell) for the cage treatment. Only the Mt Hassell population produced follicles in the bagged treatment giving a mean of 0.45 follicles per cone (Fig. 3). No differences in pollinator treatments were found between small/large or upland/lowland population groups.
Discussion
This study shows that, whilst the genetic diversity levels and reproductive output of a translocated population of B. brownii were comparable to, or surpassed those of wild reference populations, the present patterns of mating suggest that genetic diversity in the translocated population is at risk of erosion due to a significantly higher degree of selfing in this mixed mating species. The high reproductive output of the translocated population indicates that the increased inbreeding is not currently having a negative effect on seed production, though seedling fitness was not investigated and the implications for fitness in subsequent life stages are unknown.
Genetic diversity
Our results suggest that the mixing of four source populations has been beneficial in sourcing genetic diversity for the translocated population of B. brownii. The genetic diversity of the progeny of the translocated population indicated that the diversity captured in the founding population is being maintained in the F1 generation. Of the source populations, Coates et al. (2015) reported that the overall genetic diversity was highest in the upland unit and lower in the central lowland unit. The mixing of four lowland source populations has raised the diversity levels of the translocated population above any of its individual sources and closer in values to the more diverse populations of B. brownii. The diversity of founders appears to be buffering the effect of small population size within the translocation with genetic parameters aligning more closely with the most diverse B. brownii populations sampled. This is despite the translocation having only 80 reproductive individuals at the time of sampling, 5 years after its establishment. Several studies have similarly shown translocated plant populations obtaining equivalent or greater levels of diversity in comparison to wild populations, largely attributed to mixing source populations and the number (and genetic diversity) of the founders used (Ramp et al. 2006; Lloyd et al. 2012; Fant et al. 2013; Alonso et al. 2014; Zavodna et al. 2015). In contrast, other studies have found a lower genetic diversity in translocated populations relative to their source (Fant et al. 2008; Liu et al. 2008; Neale 2012). This was attributed to the use of a limited number of founders and/or a single population as a source for the translocation, contributing to the loss of genetic diversity and increased genetic divergence from the source populations (Williams and Davis 1996; Smulders et al. 2000; Liu et al. 2008; Yokogawa et al. 2013). Overall, these findings highlight the importance of sourcing seed and propagative material from numerous populations spanning the distributional range of the target species.
Amongst the wild populations, results concur with those of Coates et al. (2015) with the upland units being significantly differentiated from the central lowland units. Unlike Coates et al. (2015) genetic diversity in this study was not significantly higher in upland units, however this is likely due to the limited availability of populations to sample at the time of our study. Coates et al. (2015) sampled all known populations including those that are now extinct, where the greatest levels of genetic diversity were found. They suggested that the higher levels of genetic diversity in the upland populations, combined with significant isolation by distance among populations, likely indicate larger and more stable population sizes in these upland populations over longer timeframes. Given these historically isolated populations also occupy contrasting habitats in terms of substrate, associated vegetation and climate, and show marked differences in their mating systems (see below), our findings support the view (Coates et al. 2015) that they should be considered discrete units for management and conservation of B. brownii.
While the aims of this study focussed on assessing the success of a translocation of B. brownii, our findings have also provided the opportunity to investigate temporal genetic stability of both large and small natural populations, and the translocated population over a 7-year period. Although it covers a relatively short time frame given the longevity (40+ years) of this species, we found genetic diversity measures were comparable from adult plants in 2006 to progeny cohorts in 2013 in all populations, despite the slight (statistically insignificant) rise in the inbreeding coefficient between generations in the translocated and small lowland population. The stability of genetic diversity is persistent despite the ongoing decline in all wild populations due to the impacts of Phytophthora dieback.
The high inbreeding coefficient of the translocated population can be indicative of significant levels of selfing. However, other causes may contribute to homozygote excess such as the use of nursery grown seedlings (where early selection against selfed progeny may not occur) (Monks et al. 2021) or a Wahlund effect. The Wahlund effect can occur when genotypic proportions are calculated from heterogenous samples where individuals belonging to genetically different populations are pooled, as is the case here where we have used four source populations to establish the translocation. Both Fant et al. (2013) and Monks et al. (2021) reported high inbreeding coefficient values for translocated populations that were established from mixed sources and suggested both a Wahlund effect due to genetic sub-structuring and inbreeding as a result of small effective population size could be contributing to this result. Although it is evident that using multiple source populations to establish this translocated population has been effective in capturing levels of genetic diversity representative of the species, it is also clear that the maintenance of diversity levels obtained is highly contingent on the processes operating post establishment that influence the genetic composition of the first-generation progeny. This emphasises the need for careful consideration during site selection of factors such as number of founders, planting design and pollinator availability.
Mating systems, pollinators and reproductive output
B. brownii has been reported to have a mixed mating system with some of the highest levels of selfing recorded within the Banksia genus (Sampson et al. 1994). Our study confirms this high selfing rate and in addition, a high degree of mating pattern variation between B. brownii populations. Outcrossing rates varied from almost completely outcrossing in the large upland population to increased selfing in the lowland populations and the highest rate of selfing in the translocated population. Considerable differences in mating system parameters were evident between upland and lowland population groups, with upland populations exhibiting high outcrossing rates, minimal mating between close relatives, a difference in the order of magnitude in the number of effective pollen donors and significantly different fixation indices. These contrasting patterns appear to indicate fundamental differences in the mating system between the upland and lowland conservation units, possibly due to differing ecological and evolutionary histories for the two population groups.
Vertebrates were the most effective pollinators at all populations, although our study could not detect differences among vertebrate groups. Honeyeaters, rodents, honey possums and lizards have all been suggested as potential pollinators of B. brownii (Day et al. 1997). Differences in the proportion of flying vs non-flying vertebrates conducting pollination may affect variations in pollen dispersal patterns, with birds having higher potential mobility through sites (Carthew and Goldingay 1997; Krauss et al. 2017). Indeed, for small non-flying vertebrates that are more active in warmer conditions, such as lizards (Adolph and Porter 1993) and honey possums (Withers et al. 1989), opportunities for activity may be limited in the cool montane environment, with B. brownii flowering during autumn and winter months, and as such pollination in this habitat may predominately be carried out by birds. Nevertheless, New Holland honeyeaters (Phylidonyris novaehollandiae) were observed to be ubiquitous at all populations and are expected to be major contributors to the pollination of B. brownii. Consequently, factors influencing pollinator behaviour are likely to be influencing the patterns of mating.
The number of inflorescences produced on plants appears to be a key factor driving outcrossing rates (Fig. 2). Potential flower production is constrained by plant size and this in turn is determined by the plant’s habitat. The smaller stature imposed on the upland B. brownii by the montane environment limits the potential number of flowers produced and therefore motivates pollinators to undertake frequent inter-plant movements, resulting in the high outcrossing rates and number of effective pollen donors. In contrast, the numerous flowers produced by the much larger lowland plants allow pollinators to forage for longer periods on the same individual potentially increasing levels of self-pollination. Significantly, previous studies have reported links of mating system parameters with number of flowers per plant (Hessing 1988; Goodwillie et al. 2010) and variations in pollinator behaviour due to the spatial structure of populations (Llorens et al. 2012, 2013; Ma et al. 2019).
The low outcrossing rate in the translocated population has not however resulted in a reduction in reproductive output, with the translocated population having greater output than all other populations examined. Despite the large size of the translocated plants, which was most likely due to the high moisture levels of the south facing slope on which they were established, follicle and inflorescence production was still highest in this population even when size was accounted for (i.e. inflorescences/per unit of canopy). This finding indicates that the inbreeding detected is not imposing restrictions on seed production, although negative effects may well be realised in subsequent life-cycle stages (Husband and Schemske 1996). Indeed, it is possible that individuals produced through selfing are less likely to survive than those that are outcrossed, leaving mostly outcrossed individuals to contribute to future generations. Alternatively, species that have undergone long periods of selfing may have purged a high proportion of lethal genes and consequently exhibit lower levels of inbreeding depression (Ellstrand and Elam 1993; Sampson et al. 2016). While the relatively high levels of selfing observed in this translocated population, if sustained, would likely lead to a loss of genetic diversity over time and inbreeding depression (Frankham 2015), there are a number of factors that suggest it is probably too early to determine whether the long-term viability of this translocated population will be compromised. Importantly, maintenance of high levels of genetic diversity in the F1 seed sampled and high seed production were evident. Furthermore, many plants had not flowered at the time of sampling and it is possible as the number of flowering plants increases selfing rates may decline as pollinators move more between plants effecting outcrossing (Van Treuren et al. 1993; Karron et al. 1995). Finally, although the translocated population is more than 10 km from the nearest natural population it is possible that bird mediated pollen movement might occasionally occur over such a distance (see Byrne et al. 2007) and that this would buffer against the fitness-eroding effects of genetic drift (see for example Llorens et al. 2012, 2013).
Conclusion
Our findings indicate that establishing translocated populations from multiple source populations can conserve genetic diversity of rare and threatened plant species. However, while genetic diversity and reproductive output were comparable or higher than wild reference populations, successful outcomes in these measures can potentially mask the longer-term erosion of genetic diversity through selfing. High rates of selfing in translocated populations are of concern for the viability of population recovery success. Future study establishing the longer-term impacts/recovery of genetic diversity in populations in relation to their population size and persistence is recommended. More generally, we recommend that caution be applied when using demographic and/or genetic diversity measures on their own to assess translocation success. Patterns of mating and pollinator behaviour also need to be considered when evaluating the long-term persistence of animal pollinated translocated plant populations as plant mating systems may vary both temporally and spatially (Whitehead et al. 2018; this study). In the recently established translocation of B. brownii further studies on mating system parameters, levels of genetic diversity and reproductive output are warranted to determine whether current values for these assessments change as the population ages or whether further management intervention is needed.
Supplementary material
Supplementary material is available online.
Data availability
The data that support this study will be shared on reasonable request to the corresponding author.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This research was funded by the Australian government’s National Environmental Science Program through the Threatened Species Recovery Hub.
Acknowledgements
The authors thank Damien Rathbone, Dylan Lehman and Louisa Bell for assistance with data collection. We would also like to thank Shelley McArthur and Bronwyn MacDonald for their guidance and support in the laboratory.
References
Adolph SC, Porter WP (1993) Temperature, activity, and lizard life histories. The American Naturalist 142, 273–295.Aguilar R, Ashworth L, Galetto L, Aizen MA (2006) Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis. Ecology Letters 9, 968–980.
| Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis.Crossref | GoogleScholarGoogle Scholar |
Aguilar R, Cristóbal-Pérez EJ, Balvino-Olvera FJ, de Jesús Aguilar-Aguilar M, Aguirre-Acosta N, Ashworth L, et al. (2019) Habitat fragmentation reduces plant progeny quality: a global synthesis. Ecology Letters 22, 1163–1173.
| Habitat fragmentation reduces plant progeny quality: a global synthesis.Crossref | GoogleScholarGoogle Scholar |
Alonso MÁ, Guilló A, Pérez-Botella J, Crespo MB, Juan A (2014) Genetic assessment of population restorations of the critically endangered Silene hifacensis in the Iberian Peninsula. Journal for Nature Conservation 22, 532–538.
| Genetic assessment of population restorations of the critically endangered Silene hifacensis in the Iberian Peninsula.Crossref | GoogleScholarGoogle Scholar |
Brown AHD (1979) Enzyme polymorphism in plant populations. Theoretical Population Biology 15, 1–42.
| Enzyme polymorphism in plant populations.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Elliot CP, Yates C, Coates DJ (2007) Extensive pollen dispersal in a bird-pollinated shrub, Calothamnus quadrifidus, in a fragmented landscape. Molecular Ecology 16, 1303–1314.
| Extensive pollen dispersal in a bird-pollinated shrub, Calothamnus quadrifidus, in a fragmented landscape.Crossref | GoogleScholarGoogle Scholar |
Carthew SM, Goldingay RL (1997) Non-flying mammals as pollinators. Trends in Ecology & Evolution 12, 104–108.
| Non-flying mammals as pollinators.Crossref | GoogleScholarGoogle Scholar |
Coates DJ, Sampson JF, Yates CJ (2007) Plant mating systems and assessing population persistence in fragmented landscapes. Australian Journal of Botany 55, 239–249.
| Plant mating systems and assessing population persistence in fragmented landscapes.Crossref | GoogleScholarGoogle Scholar |
Coates DJ, McArthur SL, Byrne M (2015) Significant genetic diversity loss following pathogen driven population extinction in the rare endemic Banksia brownii (Proteaceae). Biological Conservation 192, 353–360.
| Significant genetic diversity loss following pathogen driven population extinction in the rare endemic Banksia brownii (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Collins BG, McDavitt S, Sampson JF (1993) Flowering phenology and reproductive success for an endangered species of Banksia. In ‘Proceedings of the international symposium on pollination in tropics’. pp. 130–133. (I. U. S. I.: Bangalore, India)
Commander LE, Coates DJ, Broadhurst L, Offord CA, Makinson RO, Matthes M (2018) ‘Guidelines for the translocation of threatened plants in Australia.’ 3rd edn. (Australian Network for Plant Conservation: Canberra, ACT)
Day DA, Collins BG, Rees RG (1997) Reproductive biology of the rare and endangered Banksia brownii Baxter ex R. Br. (Proteaceae). Australian Journal of Ecology 22, 307–315.
| Reproductive biology of the rare and endangered Banksia brownii Baxter ex R. Br. (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Dudash MR, Murren CJ, Carr DE (2005) Using Mimulus as a model system to understand the role of inbreeding in conservation: genetic and ecological approaches. Annals of the Missouri Botanical Garden 92, 36–51.
Eckert CG, Ozimec B, Herlihy CR, Griffin CA, Routley MB (2009) Floral morphology mediates temporal variation in the mating system of a self-compatible plant. Ecology 90, 1540–1548.
| Floral morphology mediates temporal variation in the mating system of a self-compatible plant.Crossref | GoogleScholarGoogle Scholar |
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24, 217–242.
| Population genetic consequences of small population size: implications for plant conservation.Crossref | GoogleScholarGoogle Scholar |
Fant JB, Holmstrom RM, Sirkin E, Etterson JR, Masi S (2008) Genetic structure of threatened native populations and propagules used for restoration in a clonal species, American beachgrass (Ammophila breviligulata Fern.). Restoration Ecology 16, 594–603.
| Genetic structure of threatened native populations and propagules used for restoration in a clonal species, American beachgrass (Ammophila breviligulata Fern.).Crossref | GoogleScholarGoogle Scholar |
Fant JB, Kramer A, Sirkin E, Havens K (2013) Genetics of reintroduced populations of the narrowly endemic thistle, Cirsium pitcheri (Asteraceae). Botany 91, 301–308.
| Genetics of reintroduced populations of the narrowly endemic thistle, Cirsium pitcheri (Asteraceae).Crossref | GoogleScholarGoogle Scholar |
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Molecular Ecology 24, 2610–2618.
| Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow.Crossref | GoogleScholarGoogle Scholar |
Ghazoul J (2005) Pollen and seed dispersal among dispersed plants. Biological Reviews 80, 413–443.
| Pollen and seed dispersal among dispersed plants.Crossref | GoogleScholarGoogle Scholar |
Gilfillan S, Barrett S (2005) Feather-leaved banksia (Banksia brownii) interim recovery plan 2005–2010. Department of Conservation and Land Management, Perth, WA, Australia.
Godefroid S, Piazza C, Rossi G, Buord S, Stevens A-D, Aguraiuja R, Cowell C, Weekley CW, Vogg G, Iriondo JM, Johnson I, Dixon B, Gordon D, Magnanon S, Valentin B, Bjureke K, Koopman R, Vicens M, Virevaire M, Vanderborght T (2011) How successful are plant species reintroductions? Biological Conservation 144, 672–682.
| How successful are plant species reintroductions?Crossref | GoogleScholarGoogle Scholar |
Goodwillie C, Sargent RD, Eckert CG, Elle E, Geber MA, Johnston MO, Kalisz S, Moeller DA, Ree RH, Vallejo-Marin M, Winn AA (2010) Correlated evolution of mating system and floral display traits in flowering plants and its implications for the distribution of mating system variation. New Phytologist 185, 311–321.
| Correlated evolution of mating system and floral display traits in flowering plants and its implications for the distribution of mating system variation.Crossref | GoogleScholarGoogle Scholar |
Guerrant EO (2013) The value and propriety of reintroduction as a conservation tool for rare plants. Botany 91, v–x.
| The value and propriety of reintroduction as a conservation tool for rare plants.Crossref | GoogleScholarGoogle Scholar |
Guerrant EO, Kaye TN (2007) Reintroduction of rare and endangered plants: common factors, questions and approaches. Australian Journal of Botany 55, 362–370.
| Reintroduction of rare and endangered plants: common factors, questions and approaches.Crossref | GoogleScholarGoogle Scholar |
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In ‘Plant population genetics, breeding and genetic resources’. (Eds AHD Brown, MT Clegg, AL Kahler, BS Weir) pp. 43–63. (Sinauer Press: Sunderland, MA, USA)
Hessing MB (1988) Geitonogamous pollination and its consequences in Geranium caespitosum. American Journal of Botany 75, 1324–1333.
| Geitonogamous pollination and its consequences in Geranium caespitosum.Crossref | GoogleScholarGoogle Scholar |
Husband BC, Schemske DW (1996) Evolution of the magnitude and timing of inbreeding depression in plants. Evolution 50, 54–70.
| Evolution of the magnitude and timing of inbreeding depression in plants.Crossref | GoogleScholarGoogle Scholar |
IUCN/SSC (2013) ‘Guidelines for reintroductions and other conservation translocations. Version 1.0.’ p. 57. (IUCN Species Survival Commission: Gland, Switzerland) Available at www.iucnsscrsg.org
Jachowski DS, Millspaugh JJ, Angermeier PL, Slotow R (Eds) (2016) ‘Reintroduction of fish and wildlife populations.’ (University of California Press: CA, USA)
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes 5, 187–189.
| HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness.Crossref | GoogleScholarGoogle Scholar |
Karron JD, Thumser NN, Tucker R, Hessenauer AJ (1995) The influence of population density on outcrossing rates in Mimulus ringens. Heredity 75, 175–180.
| The influence of population density on outcrossing rates in Mimulus ringens.Crossref | GoogleScholarGoogle Scholar |
Keller LF, Biebach I, Ewing SR, Hoeck PE (2012) The genetics of reintroductions: inbreeding and genetic drift. In ‘Reintroduction biology: integrating science and management’. (Eds JG Ewen, DP Armstrong, KA Parker, PJ Seddon) pp. 360–394. (Wiley: Chichester, UK)
Krauss SL, Phillips RD, Karron JD, Johnson SD, Roberts D G, Hopper SD (2017) Novel consequences of bird pollination for plant mating. Trends in Plant Science 22, 395–410.
| Novel consequences of bird pollination for plant mating.Crossref | GoogleScholarGoogle Scholar |
Lawrence BA, Kaye TN (2011) Reintroduction of Castilleja levisecta: effects of ecological similarity, source population genetics, and habitat quality. Restoration Ecology 19, 166–176.
| Reintroduction of Castilleja levisecta: effects of ecological similarity, source population genetics, and habitat quality.Crossref | GoogleScholarGoogle Scholar |
Leimu R, Mutikainen P, Koricheva J, Fischer M (2006) How general are positive relationships between plant population size, fitness and genetic variation? Journal of Ecology 94, 942–952.
| How general are positive relationships between plant population size, fitness and genetic variation?Crossref | GoogleScholarGoogle Scholar |
Liddell E, Cook CN, Sunnucks P (2020) Evaluating the use of risk assessment frameworks in the identification of population units for biodiversity conservation. Wildlife Research 47, 208–216.
| Evaluating the use of risk assessment frameworks in the identification of population units for biodiversity conservation.Crossref | GoogleScholarGoogle Scholar |
Liu M-H, Chen X-Y, Zhang X, Shen D-W (2008) A population genetic evaluation of ecological restoration with the case study on Cyclobalanopsis myrsinaefolia (Fagaceae). Plant Ecology 197, 31–41.
| A population genetic evaluation of ecological restoration with the case study on Cyclobalanopsis myrsinaefolia (Fagaceae).Crossref | GoogleScholarGoogle Scholar |
Llorens TM, Byrne M, Yates CJ, Nistelberger HM, Coates DJ (2012) Evaluating the influence of different aspects of habitat fragmentation on mating patterns and pollen dispersal in the bird-pollinated Banksia sphaerocarpa var. caesia. Molecular Ecology 21, 314–328.
| Evaluating the influence of different aspects of habitat fragmentation on mating patterns and pollen dispersal in the bird-pollinated Banksia sphaerocarpa var. caesia.Crossref | GoogleScholarGoogle Scholar |
Llorens TM, Yates CJ, Byrne M, Nistelberger HM, Williams MR, Coates DJ (2013) Complex interactions between remnant shape and the mating system strongly influence reproductive output and progeny performance in fragmented populations of a bird-pollinated shrub. Biological Conservation 164, 129–139.
| Complex interactions between remnant shape and the mating system strongly influence reproductive output and progeny performance in fragmented populations of a bird-pollinated shrub.Crossref | GoogleScholarGoogle Scholar |
Lloyd MW, Burnett RK, Engelhardt KAM, Neel MC (2012) Does genetic diversity of restored sites differ from natural sites? A comparison of Vallisneria americana (Hydrocharitaceae) populations within the Chesapeake Bay. Conservation Genetics 13, 753–765.
| Does genetic diversity of restored sites differ from natural sites? A comparison of Vallisneria americana (Hydrocharitaceae) populations within the Chesapeake Bay.Crossref | GoogleScholarGoogle Scholar |
Ma Y, Yin G, Gao J, Luo Y-B, Bai W-N (2019) Effects of distinct pollinators on the mating system and reproductive success in Incarvillea sinensis, an annual with large floral displays. Journal of Plant Ecology 12, 137–143.
| Effects of distinct pollinators on the mating system and reproductive success in Incarvillea sinensis, an annual with large floral displays.Crossref | GoogleScholarGoogle Scholar |
McArthur SL, Coates DJ (2010) Characterisation of microsatellite markers for the rare and critically endangered Banksia brownii (Proteaceae). Conservation Genetics Resources 2, 177–179.
| Characterisation of microsatellite markers for the rare and critically endangered Banksia brownii (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Menges ES (2008) Restoration demography and genetics of plants: when is a translocation successful? Australian Journal of Botany 56, 187–196.
| Restoration demography and genetics of plants: when is a translocation successful?Crossref | GoogleScholarGoogle Scholar |
Millar MA, Coates DJ, Byrne M, Krauss SL, Jonson J, Hopper SD (2019a) Assessment of genetic diversity and mating system of Acacia cyclops restoration and remnant populations. Restoration Ecology 27, 1327–1338.
| Assessment of genetic diversity and mating system of Acacia cyclops restoration and remnant populations.Crossref | GoogleScholarGoogle Scholar |
Millar MA, Anthony JM, Coates DJ, Byrne M, Krauss SL, Williams MR, Hopper SD (2019b) Genetic diversity, mating system, and reproductive output of restored Melaleuca acuminata populations are comparable to natural remnant populations. Ecological Restoration 37, 222–232.
| Genetic diversity, mating system, and reproductive output of restored Melaleuca acuminata populations are comparable to natural remnant populations.Crossref | GoogleScholarGoogle Scholar |
Millar MA, Coates DJ, Byrne M, Krauss SL, Williams MR, Jonson J, Hopper SD (2020) Pollen dispersal, pollen immigration, mating and genetic diversity in restoration of the southern plains Banksia. Biological Journal of the Linnean Society 129, 773–792.
| Pollen dispersal, pollen immigration, mating and genetic diversity in restoration of the southern plains Banksia.Crossref | GoogleScholarGoogle Scholar |
Monks L, Coates D, Bell T, Bowles ML (2012) Determining success criteria for reintroductions of threatened long-lived plants. In ‘Plant reintroduction in a changing climate’. pp. 189–208. (Island Press: Washington, DC, USA)
Monks L, Standish R, McArthur S, Dillon R, Byrne M, Coates D (2021) Genetic and mating system assessment of translocation success of the long-lived perennial shrub Lambertia orbifolia (Proteaceae). Restoration Ecology 29, e13369
| Genetic and mating system assessment of translocation success of the long-lived perennial shrub Lambertia orbifolia (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Neale JR (2012) Genetic considerations in rare plant reintroduction: practical applications (or how are we doing?). In ‘Plant reintroduction in a changing climate’. (Eds J Maschinski, KE Haskins, PH Raven) pp. 71–88. (Island Press: Washington, DC, USA)
Oostermeijer JGB, Luijten SH, Petanidou T, Kos M, Ellis-Adam AC, Den Nijs JCM (2000) Pollination in rare plants: is population size important. Det Norske Videnskaps-akademi. I. Matematisk Naturvidenskapelige Klasse, Skrifter, Ny Serie 39, 201–213.
Ottewell KM, Bickerton DC, Byrne M, Lowe AJ (2016) Bridging the gap: a genetic assessment framework for population-level threatened plant conservation prioritization and decision-making. Diversity and Distributions 22, 174–188.
| Bridging the gap: a genetic assessment framework for population-level threatened plant conservation prioritization and decision-making.Crossref | GoogleScholarGoogle Scholar |
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539.
| GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update.Crossref | GoogleScholarGoogle Scholar |
Ralls K, Ballou JD, Dudash MR, Eldridge MDB, Fenster CB, Lacy RC, Sunnucks P, Frankham R (2018) Call for a paradigm shift in the genetic management of fragmented populations. Conservation Letters 11, e12412
| Call for a paradigm shift in the genetic management of fragmented populations.Crossref | GoogleScholarGoogle Scholar |
Ramp JM, Collinge SK, Ranker TA (2006) Restoration genetics of the vernal pool endemic Lasthenia conjugens (Asteraceae). Conservation Genetics 7, 631–649.
| Restoration genetics of the vernal pool endemic Lasthenia conjugens (Asteraceae).Crossref | GoogleScholarGoogle Scholar |
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conservation Biology 17, 230–237.
| Correlation between fitness and genetic diversity.Crossref | GoogleScholarGoogle Scholar |
Ritchie AL, Krauss SL (2012) A genetic assessment of ecological restoration success in Banksia attenuata. Restoration Ecology 20, 441–449.
| A genetic assessment of ecological restoration success in Banksia attenuata.Crossref | GoogleScholarGoogle Scholar |
Ritland K (1989) Correlated matings in the partial selfer Mimulus guttatus. Evolution 43, 848–859.
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity 88, 221–228.
| Extensions of models for the estimation of mating systems using n independent loci.Crossref | GoogleScholarGoogle Scholar |
Ritland K, Jain S (1981) A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity 47, 35–52.
| A model for the estimation of outcrossing rate and gene frequencies using n independent loci.Crossref | GoogleScholarGoogle Scholar |
Rye BL, Hopper SD (1981) ‘A guide to the gazetted rare flora of Western Australia.’ Report – Department of Fisheries and Wildlife Western Australia (Australia). (Department of Fisheries and Wildlife)
Sampson JF, Collins BG, Coates DJ (1994) Mixed mating in Banksia brownii Baxter ex R. Br. (Proteaceae). Australian Journal of Botany 42, 103–111.
| Mixed mating in Banksia brownii Baxter ex R. Br. (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Sampson JF, Byrne M, Gibson N, Yates C (2016) Limiting inbreeding in disjunct and isolated populations of a woody shrub. Ecology and Evolution 6, 5867–5880.
| Limiting inbreeding in disjunct and isolated populations of a woody shrub.Crossref | GoogleScholarGoogle Scholar |
Seddon PJ (1999) Persistence without intervention: assessing success in wildlife reintroductions. Trends in Ecology & Evolution 14, 503
| Persistence without intervention: assessing success in wildlife reintroductions.Crossref | GoogleScholarGoogle Scholar |
Seddon PJ, Armstrong DP, Maloney RF (2007) Developing the science of reintroduction biology. Conservation Biology 21, 303–312.
| Developing the science of reintroduction biology.Crossref | GoogleScholarGoogle Scholar |
Silcock JL, Simmons CL, Monks L, Dillon R, Reiter N, Jusaitis M, et al. (2019) Threatened plant translocation in Australia: a review. Biological Conservation 236, 211–222.
| Threatened plant translocation in Australia: a review.Crossref | GoogleScholarGoogle Scholar |
Smulders MJM, van der Schoot J, Geerts RHEM, Antonisse-de Jong AG, Korevaar H, van der Werf A, Vosman B (2000) Genetic diversity and the reintroduction of meadow species. Plant Biology 2, 447–454.
| Genetic diversity and the reintroduction of meadow species.Crossref | GoogleScholarGoogle Scholar |
St. Clair AB, Dunwiddie PW, Fant JB, Kaye TN, Kramer AT (2020) Mixing source populations increases genetic diversity of restored rare plant populations. Restoration Ecology 28, 583–593.
| Mixing source populations increases genetic diversity of restored rare plant populations.Crossref | GoogleScholarGoogle Scholar |
Van Rossum F, Hardy OJ, Le Pajolec S, Raspé O (2020) Genetic monitoring of translocated plant populations in practice. Molecular Ecology 29, 4040–4058.
| Genetic monitoring of translocated plant populations in practice.Crossref | GoogleScholarGoogle Scholar |
Van Treuren R, Bulsma R, Ouborg NJ, Van Delden W (1993) The effects of population size and plant density on outcrossing rates in locally endangered Salvia pratensis. Evolution 47, 1094–1104.
| The effects of population size and plant density on outcrossing rates in locally endangered Salvia pratensis.Crossref | GoogleScholarGoogle Scholar |
Weeks AR, Sgro CM, Young AG, Frankham R, Mitchell NJ, Miller KA, et al. (2011) Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evolutionary Applications 4, 709–725.
| Assessing the benefits and risks of translocations in changing environments: a genetic perspective.Crossref | GoogleScholarGoogle Scholar |
Weeks AR, Stoklosa J, Hoffmann AA (2016) Conservation of genetic uniqueness of populations may increase extinction likelihood of endangered species: the case of Australian mammals. Frontiers in Zoology 13, 31
| Conservation of genetic uniqueness of populations may increase extinction likelihood of endangered species: the case of Australian mammals.Crossref | GoogleScholarGoogle Scholar |
Wendelberger KS, Fellows MQN, Maschinski J (2008) Rescue and restoration: experimental translocation of Amorpha herbacea Walter var. crenulata (Rybd.) Isley into a novel urban habitat. Restoration Ecology 16, 542–552.
| Rescue and restoration: experimental translocation of Amorpha herbacea Walter var. crenulata (Rybd.) Isley into a novel urban habitat.Crossref | GoogleScholarGoogle Scholar |
Whitehead MR, Lanfear R, Mitchell RJ, Karron JD (2018) Plant mating systems often vary widely among populations. Frontiers in Ecology and Evolution 6, 38
| Plant mating systems often vary widely among populations.Crossref | GoogleScholarGoogle Scholar |
Williams SL, Davis CA (1996) Population genetic analyses of transplanted eelgrass (Zostera marina) beds reveal reduced genetic diversity in southern California. Restoration Ecology 4, 163–180.
| Population genetic analyses of transplanted eelgrass (Zostera marina) beds reveal reduced genetic diversity in southern California.Crossref | GoogleScholarGoogle Scholar |
Withers PC, Richardson KC, Wooller RD (1989) Metabolic physiology of euthermic and torpid honey possums, Tarsipes rostratus. Australian Journal of Zoology 37, 685–693.
| Metabolic physiology of euthermic and torpid honey possums, Tarsipes rostratus.Crossref | GoogleScholarGoogle Scholar |
Yokogawa M, Kaneko S, Takahashi Y, Isagi Y (2013) Genetic consequences of rapid population decline and restoration of the critically endangered herb Polemonium kiushianum. Biological Conservation 157, 401–408.
| Genetic consequences of rapid population decline and restoration of the critically endangered herb Polemonium kiushianum.Crossref | GoogleScholarGoogle Scholar |
Zavodna M, Abdelkrim J, Pellissier V, Machon N (2015) A long-term genetic study reveals complex population dynamics of multiple-source plant reintroductions. Biological Conservation 192, 1–9.
| A long-term genetic study reveals complex population dynamics of multiple-source plant reintroductions.Crossref | GoogleScholarGoogle Scholar |