Determining the generation length and level of morphological and genetic differentiation in the Data Deficient glossy grass skink (Pseudemoia rawlinsoni)
Lucy Wotherspoon A , Margaret L. Haines A B , Jules E. Farquhar A # and David G. Chapple A # *
A # *
A
B
Handling Editor: Dan Lunney
Abstract
Human activities are having a significant impact on biodiversity worldwide, to the extent that we are in the midst of the sixth mass extinction event. Although a substantial proportion of species globally have an elevated risk of extinction, some species are poorly known and there is insufficient information available to adequately assess their risk of extinction.
One such species is the glossy grass skink (Pseudemoia rawlinsoni), a widespread but enigmatic lizard species in south-eastern Australia. In order to improve our knowledge of its life history, and particularly its generation length, we examined museum specimens collected from across the range of the species, supplemented with measurements from field-caught individuals.
We estimated that the species reaches sexual maturity in 3 years, at approximately 40 mm snout–vent length. Its generation length was estimated as 5 years. Sexual dimorphism was evident, and female body size was positively related to litter size. Although there was no evidence for substantial variation in morphology across the range of the glossy grass skink, a phylogeographic analysis using mitochondrial DNA sequence data (ND4) revealed the presence of seven genetic sublineages (up to 5.1% genetic divergence) within the species.
The glossy grass skink appears to be a single, but widespread and genetically variable, species.
Our study demonstrates how a targeted, multifaceted study can be effective at rapidly gathering data that can be used to contribute vital information to the assessment of extinction risk in Data Deficient species.
Keywords: Australia, extinction risk, IUCN Red List, life history, lizard, phylogeography, reproductive ecology, Scincidae.
Introduction
Biodiversity is currently under increasing threat from human-mediated impacts such as habitat loss and fragmentation, invasive species, overexploitation and climate change (Doherty et al. 2016; Pecl et al. 2017; IPBES 2019). The cumulative impacts of these processes have ushered in a sixth mass extinction event, which is predicted to intensify in the future (Barnosky et al. 2011; Ceballos et al. 2015, 2020). Predicting the impact of these threats on a species requires detailed information about the species biology, ecology, habitat requirements and likely resilience to each threat (Pimm et al. 2014; Johnson et al. 2017). Under the International Union for Conservation of Nature (IUCN) Red List of Threatened Species, the extinction risk of species is estimated using a range of parameters, including generation length, population size, geographic range size, number of populations/subpopulations, and population connectivity/fragmentation (Mace et al. 2008; IUCN Standards and Petitions Committee 2022). However, where there is insufficient knowledge of these key parameters, it may not be possible to conduct an assessment of the conservation status of the species (IUCN Standards and Petitions Committee 2022).
Species may be classified as Data Deficient due to a lack of knowledge regarding their distribution, biology and ecology (e.g. life history, generation length, habitat requirements), threats to the species, and/or taxonomic status (Bland and Böhm 2016; IUCN Standards and Petitions Committee 2022; Wotherspoon et al. 2024). Without this critical knowledge, it is difficult to determine the threats that the species faces, assess their capacity to withstand and recover from any potential threats, and/or evaluate whether it represents a valid taxonomic entity. Although Data Deficient species are often overlooked by, or are a low priority for, conservation managers (Bland et al. 2017), recent studies have highlighted that they are more likely to be threatened than already assessed species (Gumbs et al. 2020; Caetano et al. 2022; Graham et al. 2023). Thus, targeted studies are required to obtain the information necessary to resolve the knowledge gaps, allowing a conservation assessment to be conducted for the species.
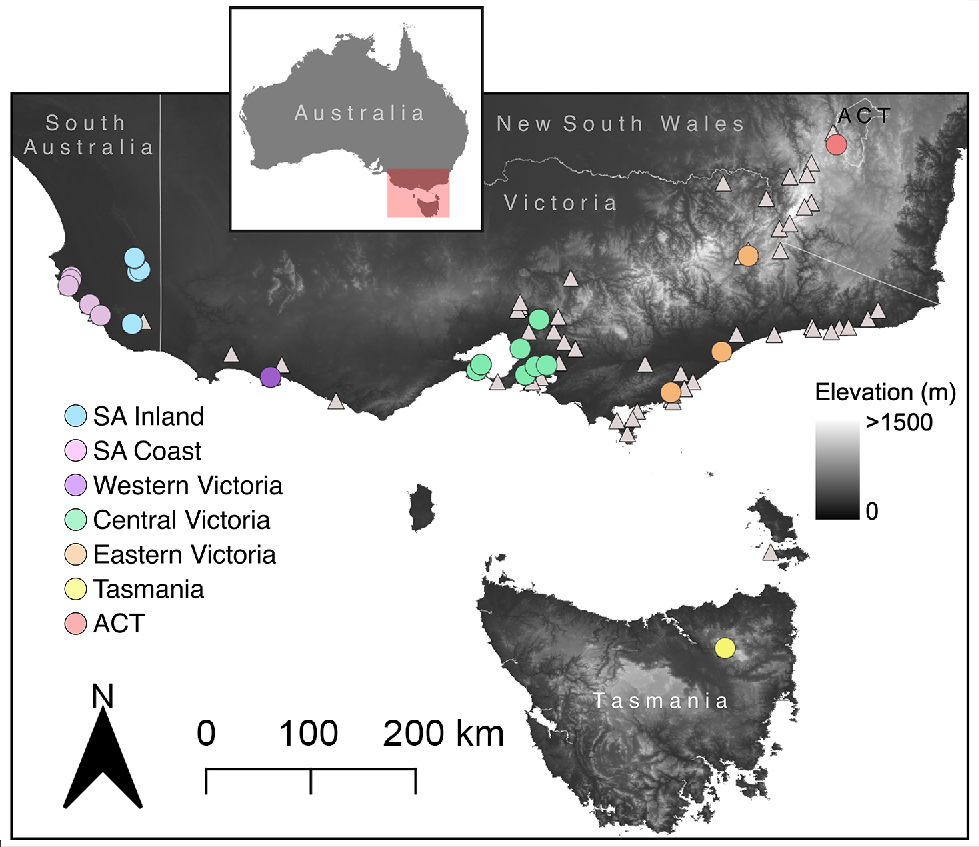
The glossy grass skink (Pseudemoia rawlinsoni) is an example of a Data Deficient lizard species that has a widespread, but disjunct, distribution in south-eastern Australia (Fig. 1), where the majority of the Australian human population resides (Gillespie et al. 2018; Chapple et al. 2019; Wilson and Swan 2021). Described in 1988 (Hutchinson and Donnellan 1988), it is an enigmatic species that inhabits moist, densely vegetated wetland habitats (Gillespie et al. 2018; Wilson and Swan 2021). State-based conservation listings in the core of the species’ range (Victoria: Endangered, South Australia: Vulnerable, Tasmania: Rare) hint at a potential elevated extinction risk for the species (Chapple et al. 2019). However, the glossy grass skink is currently listed as Data Deficient on the IUCN Red List (Gillespie et al. 2018). This is because, prior to the recent study by Farquhar et al. (2023), across the broader range of the species there was uncertainty regarding its distribution (Area of Occupancy), and before it was confirmed by Farquhar et al. (2024), it was unknown whether the species was under pressure from infrastructure development and agriculture. However, the species’ life history and generation length, number of populations/subpopulations, and its sensitivity to the impacts of invasive species are still largely unknown (Gillespie et al. 2018).
Locations of glossy grass skink (Pseudemoia rawlinsoni) specimens. Coloured circles are locations with both morphological and genetic data, with colours corresponding to genetic lineages. White triangles are locations with only morphology data.
Here we complete a targeted, multifaceted study to rapidly obtain key aspects of the information needed to conduct an IUCN Red List assessment for the glossy grass skink. Using a combination of museum and field-caught specimens, we investigate the life history and reproductive ecology of the species. Specifically, we aim to estimate the generation length of the gloss grass skink, as it is a core component of most IUCN Red List criteria, and a critical determinant of the capacity of a species to respond to, and recover from, threatening processes (Mace et al. 2008; IUCN Standards and Petitions Committee 2022). Specifically, generation length is required to determine the timeframe over which a reduction in population size should be assessed (Criterion A; 3 generations or 10 years), decline in the number of mature individuals (Criterion C1; 1, 2, or 3 generations for Critically Endangered, Endangered, and Vulnerable, respectively), and the timeframe for quantitative analyses of extinction probability (Criterion E; 3 or 5 generations for Critically Endangered and Endangered, respectively) (IUCN Standards and Petitions Committee 2022). In addition, we use mitochondrial DNA sequence data (ND4) to conduct a phylogeographic study of the gloss grass skink. This molecular study will provide some preliminary, broad scale information on the level of genetic structuring among populations across the range of the species, and assist with the determination of the number of subpopulations in the species.
Methods
Morphology, life history and reproductive ecology of the glossy grass skink
To quantify the external morphology of the glossy grass skink, we obtained data from both preserved museum specimens and field caught specimens. Firstly, we examined all preserved glossy grass skink specimens from Museums Victoria (n = 88) and the South Australian Museum (n = 18) (Fig. 1). Digital callipers (Mitutoyo 500–763–20 8″/200 mm Coolant Proof Digimatic Calipers; West Heidelberg, Australia) were used to obtain the morphological measurements (in mm) of snout–vent length (SVL), snout-axilla length (SAL), axilla–groin length (AGL), interlimb length (ILL), tail length (TL), tail width (TW), body width (BW), body height (BH), pelvic width (PW), pelvic height (PH), total front limb length (TFL), upper front limb (UFL), lower front limb (LFL), front foot (FF), total hind limb length (THL), upper hind limb (UHL), lower hind limb (LHL), hind foot (HF), head width (HW), head length (HL), head depth (HD), snout length (SL) and eye diameter (ED) (see Supplementary Table S1 for definitions of each trait; Greer 1982; Barter et al. 2022). In addition, we conducted scale counts (as outlined in Taylor 1935) of midbody scale rows, paravertebral scales, subdigital lamellae, nuchals, presuboculars, supraciliaries, supralabials and infralabials (Supplementary Fig. S1).
To examine sexual maturity, and litter size in females, in the preserved specimens, we followed the methods of Barter et al. (2022). Briefly, we made a small ventral incision along the left side of the specimen to determine the sex and sexual maturity of each individual. If an existing incision was already present, that was used instead. We considered male specimens to be sexually mature if they had fully developed testes and the ductus deferens appeared to be rough (rather than smooth, as found in juveniles) (Barter et al. 2022). Females were assessed for sexual maturity based on the presence of a folded oviduct extending cranially from the ovary (Barter et al. 2022). In juvenile females this portion of the oviduct is smooth and not folded (Barter et al. 2022). For specimens that appeared to be gravid upon external examination, the incision was made down the midline to count the number of eggs. The reproductive condition (gravid or non-gravid) was recorded for each sexually mature female, along with the litter size. Only adult specimens were used to calculate trait means.
Secondly, we measured field-caught glossy grass skinks collected at six sites across Victoria, Australia, as part of a related study (Farquhar et al. 2024). The fieldwork was conducted between October 2021 and April 2022, and COVID-19 related border closures and travel restrictions prevented us sampling populations from outside of Victoria. We measured a total of 70 specimens across these sites, shown here with latitude and longitude: Port Fairy (−38.346409, 142.242085; eight specimens), Queenscliff (−38.268226, 144.628523; 28 specimens), Swan Bay (−38.201634, 144.676130; 20 specimens), Yering (−37.681576, 145.349988; four specimens), Tooradin (−38.211158, 145.427389; one specimen) and Warringine Park (−38.325837, 145.191561; nine specimens) (Fig. 1). Snout–vent length and TL were measured using a ruler (in mm), and digital callipers (Mitutoyo 500–763–20 8″/200 mm Coolant Proof Digimatic Calipers) were used to measure (in mm) HL, HW and HD. Sex was determined by hemipenal eversion in males, or lack thereof in females. We took high-resolution photographs (Canon 5D Mark IV digital camera with a Canon 100 mm f/2.8 L macro lens) of each individual and used a combination of unique external features (scars, missing digits, aberrations in pattern, proportion of tail that was regenerated) and sex to track the identity of each individual over time, and to ensure that we did not measure the same individual twice.
All data analyses were conducted in R (Ver. 4.0.4; R Core Team 2021). To create a summary of all glossy grass skink traits, means, standard errors and ranges were calculated for each morphological measurement. To determine the size at maturity, we recorded the smallest sexually mature individual of each sex. To estimate the age at maturity we created a histogram of SVL of museum and field specimens. Fung and Waples (2017) recommended that, when estimating generation length for reptiles, the information required to develop detailed life history tables is best achieved using the following formula: generation length = age at first reproduction + z × (maximum age − age at first reproduction), where z represents a constant dependent on survivorship and relative fecundity of individuals in the population (Fung and Waples 2017). We have established a generalised z value of 0.5 due to increased fecundity with size in lizards (Olsson and Shine 1996; Du et al. 2005; Meiri et al. 2020). Although lizards grow rapidly and indeterminately, their growth rate slows following maturity (Olsson and Shine 1996; Laver et al. 2012). However, such increases in fecundity with age are often offset by gradual senescence (Patnaik 1994) and increased mortality with age. Thus, we consider this approach to be appropriate for estimating the generation length of the glossy grass skink.
All body measurements were adjusted to SVL using an allometric growth model (package GroupStruct; Chan and Grismer 2022). We used a Wilcoxon rank sum test to examine sexual dimorphism for each adjusted morphological measurement. Field and preserved specimen measurements were combined in these analyses because the amount of shrinkage endured by preserved specimens will not significantly impact body measurements (Vervust et al. 2009; Maayan et al. 2022). Additionally, we compared adjusted morphological measurements amongst the populations determined by the genetic groupings using a Kruskal–Wallis rank sum test. To investigate significant differences, we conducted a Dunn test (package dunn.test) with a Bonferroni adjustment. Finally, a linear regression analysis was conducted to determine the relationship between SVL and litter size of preserved specimens.
Assessing the presence of genetic substructuring across the range of the glossy grass skink
We obtained 46 tissue samples of the glossy grass skink across its range in south-eastern Australia (South Australian Museum, n = 17; wild caught samples from the 2021/2022 field season, n = 29; Table 1, Fig. 1). Genomic DNA was extracted from glossy grass skink liver or tail-tip tissue using a Qiagen DNeasy Blood and Tissue Extraction Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. For all samples, we amplified the mitochondrial DNA (mtDNA) fragment NADH subunit 4 (ND4; ~700 bp) using the primers ND4 and tRNA-leu, and the PCR protocols as outlined in Haines et al. (2014). We used the ND4 mtDNA gene provides similar phylogenetic resolution to other mtDNA coding regions (e.g. Chapple et al. 2009), and it was the mtDNA gene that has been most widely used for Pseudemoia, allowing us to place our glossy grass skink sequences into context with the broader genus (Haines et al. 2014). Additionally, Haines et al. (2014), using ND4 and five nuclear genes, provided evidence that the glossy grass skink is a distinct species, with no evidence of hybridisation or introgression with other Pseudemoia species. Consequently, we focused on a single mtDNA gene in the present study to document the level of intraspecific genetic divergence among populations/regions in the glossy grass skink.
| Species | Region | Locality | Tissue ID | Specimen ID | GenBank accession | |
|---|---|---|---|---|---|---|
| Pseudemoia rawlinsoni | Australian Capital Territory | Mt Bimberi | ABTC40954 | SAMR23098 | OR469914 | |
| Eastern Victoria | Woodside | ABTC04064 | – | KM263322 | ||
| Hollands Landing | NMVZ73965 | – | OR469929 | |||
| Benambra | NMVZ73970 | – | OR469928 | |||
| Central Victoria | Warringine Park | NMVZ73937 | – | OR469930 | ||
| Spadonis Nature Reserve | NMVZ73941 | – | OR469931 | |||
| Warringine Park | NMVZ73944 | – | OR469932 | |||
| Swan Bay | NMVZ73945 | – | OR469933 | |||
| Spadonis Nature Reserve | NMVZ73946 | – | OR469934 | |||
| Spadonis Nature Reserve | NMVZ73949 | – | OR469935 | |||
| Warringine Park | NMVZ73953 | – | OR469936 | |||
| Swan Bay | NMVZ73955 | – | OR469915 | |||
| Swan Bay | NMVZ73956 | – | OR469937 | |||
| Swan Bay | NMVZ73957 | – | OR469941 | |||
| Warringine Park | NMVZ73958 | – | OR469944 | |||
| Waterways | NMVZ73971 | – | OR469918 | |||
| Warringine Park | NMVZ73973 | – | OR469943 | |||
| Warneet | NMVZ73974 | – | OR469916 | |||
| Warneet | NMVZ73975 | – | OR469945 | |||
| Queenscliff | NMVZ73976 | – | OR469946 | |||
| Queenscliff | NMVZ73977 | – | OR469938 | |||
| Queenscliff | NMVZ73978 | – | OR469947 | |||
| Warringine Park | NMVZ73979 | – | OR469948 | |||
| Warringine Park | NMVZ73980 | – | OR469939 | |||
| Tooradin Inlet | NMVZ73981 | – | OR469917 | |||
| Queenscliff | NMVZ73982 | – | OR469942 | |||
| Queenscliff | NMVZ73983 | – | OR469940 | |||
| Western Victoria | Port Fairy | NMVZ73954 | – | OR469949 | ||
| Port Fairy | NMVZ73963 | – | OR469919 | |||
| Port Fairy | NMVZ73966 | – | OR469950 | |||
| Port Fairy | NMVZ73969 | – | OR469927 | |||
| Tasmania | 4.2 km SE of Burns Creek, North Esk River | ABTC57877 | SAMR44322 | KM263321 | ||
| Inland South Australia | Lake Ormerod | ABTC156252 | SAMR59775 | OR469911 | ||
| Lake Ormerod | ABTC156253 | SAMR59776 | OR469913 | |||
| Lake Ormerod | ABTC68946 | SAMR53512 | OR469912 | |||
| 5.4 km SSE of Glencoe | ABTC37606 | SAMR49405 | OR469908 | |||
| Bool Lagoon Game Reserve | ABTC54784 | SAMR23098 | OR469909 | |||
| 4.1 km N of Bool Lagoon | ABTC68787 | SAMR52584 | OR469910 | |||
| 7 km ESE of Kangaroo Hill | ABTC94787 | SAMR53753 | OR469923 | |||
| Coastal South Australia | Silky Tea Tree Swamp | ABTC106240 | SAMR57100 | OR469924 | ||
| 20 WNW of Millicent Airport | ABTC37531 | SAMR49585 | OR469925 | |||
| 20 WNW of Millicent Airport | ABTC37542 | SAMR49589 | OR469926 | |||
| 20 WNW of Millicent Airport | ABTC37543 | SAMR49590 | OR469922 | |||
| 20 WNW of Millicent Airport | ABTC37727 | SAMR49594 | OR469920 | |||
| 9 km SW of Millicent | ABTC70044 | SAMR53777 | KM263323 | |||
| 9 km SSE of Kangaroo Hill | ABTC94794 | SAMR53758 | OR469921 | |||
| Outgroups | ||||||
| Carinascincus metallicus | NA | Wilsons Promontory, Refuge Cove, Victoria | NMVZ21551 | NMVD75207 | KM263269 | |
| Pseudemoia baudini | NA | Wanna, South Australia | ABTC15402 | NMVD60966 | KM263201 | |
| Pseudemoia cryodroma | NA | Mt Baw Baw, Victoria | ABTC14601 | NMVD59893 | KM263204 | |
| Pseudemoia entrecasteauxii | NA | Mt Kosciusko, New South Wales | ABTC11167 | NMVD59933 | KM263231 | |
| Pseudemoia pagenstecheri | NA | Lake Corangamite, Victoria | NMVZ23650 | – | KM263319 | |
| Pseudemoia spenceri | NA | Mt Baw Baw, Victoria | NMVZ19287 | NMVD74853 | KM263324 |
Specimen codes: SAMR, South Australian Museum; NMVD, Museums Victoria, Melbourne. Tissue ID collection codes: ABTC, South Australian Museum Australian Biological Tissue Collection; NMVZ, Museums Victoria Frozen Tissue Collection.
Purifications and sequencing were performed by Macrogen, Inc. (Seoul, South Korea). The sequences were aligned and edited in BioEdit v7.2.5 (Hall 1999). We checked for stop codons using MUSCLE within Geneious v.10.2.6 (Biomatters, Auckland, New Zealand) and none were observed. Three glossy grass skink samples were already sequenced using the same protocol (GenBank numbers: KM263323, KM263322, KM263321; Haines et al. 2014), and were used in our study. The ND4 sequences obtained in our study were deposited in GenBank (Table 1). Outgroup sequences for the metallic skink (Carinascincus metallicus) and other members of the Pseudemoia genus were obtained from Haines et al. (2014) and GenBank (Table 1).
We used jModelTest 2.1.10 (Darriba et al. 2012) to identify the most appropriate model of sequence evolution based on the Akaike Information Criterion (AIC). We conducted Maximum Likelihood analysis using a RAxML GTR + I + G model with 100 bootstraps (Stamatakis 2014), and a Bayesian analysis using MrBayes 3.2 (Huelsenbeck and Ronquist 2001), in Geneious v.10.2.6 (Biomatters, Auckland, New Zealand) to determine the genetic structuring of glossy grass skink populations across south-eastern Australia. A genetic distancing estimation was calculated using MEGA v.11.0.13 (Tamura et al. 2004, 2021) to determine the approximate percentage of genetic variation and time of separation amongst the populations.
Results
Morphology, life history and reproductive ecology of the glossy grass skink
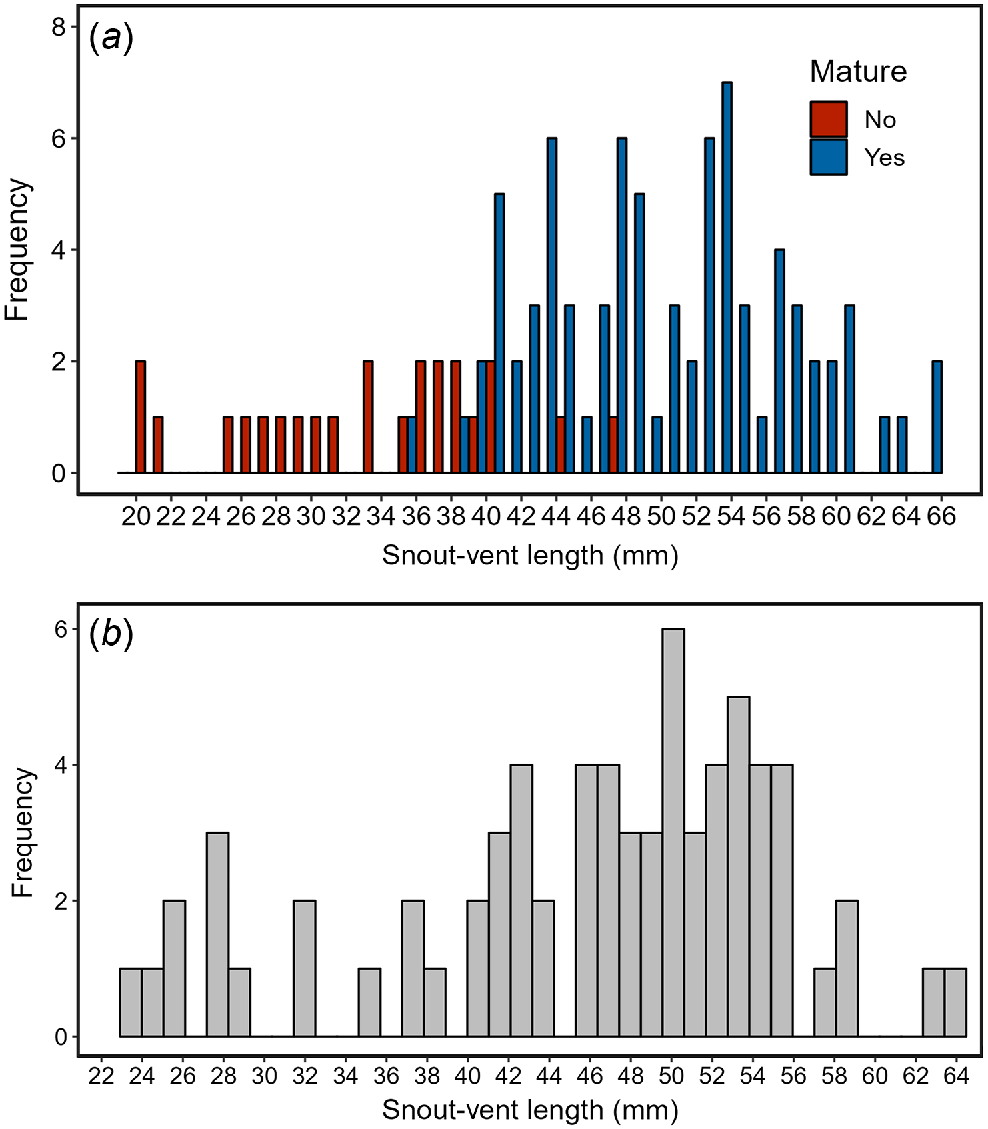
Our visual examination of the gonads of museum specimens indicated that the smallest sexually mature individual was approximately 40 mm (females: 39.9 mm, males: 38.3 mm [with an outlier individual mature at 35.7 mm]; Fig. 2a). Using this estimate for the size at maturity, inspection of the SVL of field caught glossy grass skinks in Victoria suggests that individuals may reach between 23 and 30 mm SVL during their first season, 32 and 39 mm SVL during their second season, and sexual maturity (>40 mm SVL) during their third season (Fig. 2b). Assuming an average lifespan of ~7 years for similar-sized skinks in south-eastern Australia (Greer 2022), we used the generation length formula developed by Fung and Waples (2017) to determine the generation length of the glossy grass skink as approximately 5 years (3 + 0.5 × (7 − 3) = 5).
A histogram of the (a) snout–vent length (mm) of all mature (blue) and immature (red) glossy grass skink preserved specimens (n = 103) examined in this study, and (b) snout–vent length (mm) of all field caught glossy grass skinks (n = 70) in Victoria, Australia.
Sexual dimorphism is evident in the glossy grass skink, with males having larger heads (head length, head width, snout length, eye diameter), longer axilla–groin lengths, and longer legs (total front limb length, upper front limb length, lower front limb length, front foot, total hind limb length, upper hind limb length, lower hind limb length, hind foot; Table 2). In contrast, females have larger bodies (paravertebral scale count, interlimb length, body width, body height), longer snout–axilla lengths, and greater tail widths (Table 2). All other body measurements and scale counts were not sexually dimorphic in the glossy grass skink (Table 2).
| Female | Male | Overall | Sexual Dimorphism | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | N | Mean ± s.e. | Range | N | Mean ± s.e. | Range | N | Mean ± s.e. | Range | ||
| Snout–vent length | 78 | 51.0 ± 0.75 | 40.0–65.2 | 56 | 49.3 ± 0.68 | 40.7–60.1 | 136 | 50.1 ± 0.53 | 40–65.2 | 0.129 | |
| Snout–axilla length | 42 | 28.0 ± 0.88 | 17.2–39.5 | 36 | 25.1 ± 0.82 | 16.8–32.9 | 80 | 26.5 ± 0.61 | 16.8–39.5 | <0.001 | |
| Axilla–groin distance | 42 | 18.9 ± 0.98 | 13.8–38.3 | 36 | 20.7 ± 0.86 | 15.6–34.3 | 80 | 19.7 ± 0.65 | 13.8–38.3 | <0.001 | |
| Interlimb length | 42 | 31.5 ± 0.83 | 22.9–42.0 | 36 | 28.1 ± 0.68 | 17.7–35.2 | 80 | 29.8 ± 0.58 | 17.7–42.0 | <0.001 | |
| Tail length | 26 | 79.7 ± 2.01 | 66.4–105.0 | 21 | 79.3 ± 3.08 | 43.0–104.0 | 47 | 79.5 ± 1.75 | 43.0–105 | 0.149 | |
| Tail width | 42 | 5.1 ± 0.11 | 3.8–6.6 | 36 | 4.7 ± 0.10 | 3.2–5.8 | 80 | 4.8 ± 0.08 | 3.2–6.6 | 0.018 | |
| Body width | 42 | 8.7 ± 0.24 | 5.9–12.1 | 36 | 7.4 ± 0.19 | 5.5–9.5 | 80 | 8.1 ± 0.17 | 5.4–12.1 | <0.001 | |
| Body height | 42 | 6.1 ± 0.21 | 3.6–8.6 | 36 | 4.9 ± 0.14 | 3.3–6.4 | 80 | 5.2 ± 0.14 | 3.1–8.6 | 0.001 | |
| Pelvic width | 42 | 4.4 ± 0.12 | 3.1–6.2 | 36 | 4.5 ± 0.11 | 2.8–5.5 | 80 | 4.4 ± 0.08 | 2.8–6.2 | 0.069 | |
| Pelvic height | 42 | 4.8 ± 0.11 | 3.2–6.2 | 36 | 4.8 ± 0.12 | 3.5–6.0 | 80 | 4.8 ± 0.08 | 3.2–6.2 | 0.404 | |
| Total front limb length | 42 | 11.5 ± 0.20 | 9.3–15.9 | 36 | 12.4 ± 0.21 | 9.2–14.6 | 80 | 11.9 ± 0.15 | 9.2–15.9 | <0.001 | |
| Upper front limb | 42 | 4.1 ± 0.08 | 3.3–5.4 | 36 | 4.3 ± 0.09 | 2.7–5.4 | 80 | 4.2 ± 0.06 | 2.7–5.4 | 0.001 | |
| Lower front limb | 42 | 3.9 ± 0.08 | 3.2–5.3 | 36 | 4.0 ± 0.08 | 3.1–5.1 | 80 | 4.0 ± 0.05 | 3.1–5.3 | 0.005 | |
| Front foot | 42 | 5.0 ± 0.12 | 3.3–7.4 | 36 | 5.5 ± 0.12 | 4.2–7.0 | 80 | 5.2 ± 0.09 | 3.3–7.4 | <0.001 | |
| Total hind limb length | 42 | 17.5 ± 0.34 | 13.8–22.3 | 36 | 18.7 ± 0.31 | 15.3–23.1 | 80 | 18.0 ± 0.24 | 13.8–23.1 | <0.001 | |
| Upper hind limb | 42 | 5.9 ± 0.14 | 4.5–7.9 | 36 | 6.2 ± 0.13 | 5.1–8.2 | 80 | 6.0 ± 0.09 | 4.5–8.2 | <0.001 | |
| Lower hind limb | 42 | 5.3 ± 0.11 | 4.1–7.1 | 36 | 5.7 ± 0.11 | 4.2–6.8 | 80 | 5.5 ± 0.08 | 4.1–7.1 | <0.001 | |
| Hind foot | 42 | 8.8 ± 0.19 | 6.4–12.0 | 36 | 9.6 ± 0.17 | 7.6–11.9 | 80 | 9.1 ± 0.13 | 6.4–12.0 | <0.001 | |
| Head width | 78 | 6.1 ± 0.07 | 5.2–8.3 | 56 | 6.4 ± 0.07 | 5.4–7.7 | 136 | 6.2 ± 0.05 | 5.2–8.3 | <0.001 | |
| Head length | 78 | 8.6 ± 0.10 | 5.0–10.9 | 56 | 9.4 ± 0.11 | 7.6–11.3 | 136 | 8.9 ± 0.08 | 5.0–11.3 | <0.001 | |
| Head depth | 78 | 4.6 ± 0.07 | 3.4–6.3 | 56 | 4.6 ± 0.07 | 3.4–5.6 | 136 | 4.6 ± 0.05 | 3.4–6.3 | 0.129 | |
| Snout length | 42 | 5.4 ± 0.08 | 4.5–6.9 | 36 | 5.8 ± 0.08 | 5.0–6.9 | 80 | 5.6 ± 0.06 | 4.5–6.9 | <0.001 | |
| Eye diameter | 42 | 1.1 ± 0.02 | 0.9–1.3 | 36 | 1.1 ± 0.02 | 0.9–1.3 | 80 | 1.1 ± 0.01 | 0.9–1.3 | 0.007 | |
| Midbody scales | 42 | 26.8 ± 0.16 | 26–30 | 36 | 26.7 ± 0.18 | 25–30 | 80 | 26.8 ± 0.12 | 25–30 | 0.236 | |
| Paravertebral scales | 42 | 58.1 ± 0.30 | 54–62 | 36 | 56.4 ± 0.33 | 53–59 | 80 | 57.3 ± 0.24 | 53–62 | <0.001 | |
| Subdigital lamellae | 42 | 19.2 ± 0.21 | 17–23 | 36 | 20.1 ± 0.24 | 17–23 | 80 | 19.6 ± 0.16 | 17–23 | 0.166 | |
| Nuchals | 42 | 4.6 ± 0.22 | 2–8 | 36 | 4.3 ± 0.24 | 2–8 | 80 | 4.5 ± 0.16 | 2–8 | 0.329 | |
| Presubocular | 42 | 2.2 ± 0.06 | 1–3 | 36 | 2.2 ± 0.08 | 1–3 | 80 | 2.2 ± 0.05 | 1–3 | 0.996 | |
| Supraciliaries | 42 | 6.8 ± 0.07 | 6–8 | 36 | 6.8 ± 0.08 | 6–8 | 80 | 6.8 ± 0.05 | 6–8 | 0.433 | |
| Supralabials | 42 | 7.0 ± 0.02 | 7–8 | 36 | 7.1 ± 0.04 | 7–8 | 80 | 7.1 ± 0.03 | 7–8 | 0.162 | |
| Infralabials | 42 | 7.0 ± 0.04 | 6–8 | 36 | 7.1 ± 0.05 | 6–8 | 80 | 7.1 ± 0.03 | 6–8 | 0.203 | |
The sexual dimorphism column presents the results of the size-corrected Wilcoxon rank sum exact tests for sexual dimorphism. The sex of two museum specimens was unknown. Significant results are presented in bold.
The mean litter size of glossy grass skink museum specimens was 4.65 ± 0.43 (range 1–9, n = 20). There was a positive association between female SVL and litter size (estimate = 2.58 ± 0.51, t-value = 5.10, P < 0.001; Fig. S2).
Assessing the presence of genetic substructuring across the range of the glossy grass skink
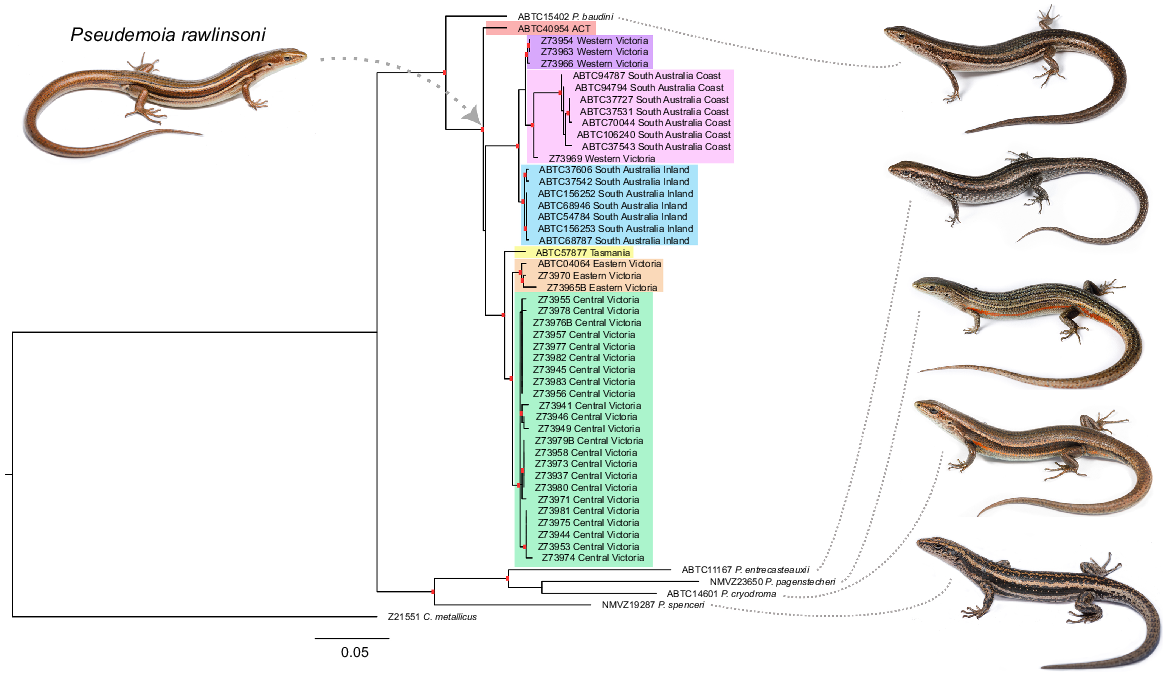
Our phylogenetic analysis supports the glossy grass skink being a single species, but with seven distinct genetic lineages across south-eastern Australia: Australian Capital Territory, Eastern Victoria, Tasmania, Central Victoria, Western Victoria, inland South Australia, and coastal South Australia (Fig. 3). The degree of genetic distance amongst lineages varied between 0.92% and 5.13% (Table 3). The seven genetic lineages of the glossy grass skink did not differ in a range of body measurements (SVL, ILL, PW, PH, LFL, THL, UHL, LHL, SL, ED) or scale counts (midbody scales, subdigital lamellae) (Tables S2 and S3). Although significant differences were found among the lineages for a range of body measurements (SAL, AGL, HL, HW, HL, TW, BW, BH, TFL, UFL, FF, HF) and scale counts (nuchals, paravertebral, presuboculars, supraciliaries, supralabials, infralabials), no consistent differences were observed (Tables S2–S22).
Maximum likelihood phylogeny for the glossy grass skink (Pseudemoia rawlinsoni) based on mitochondrial DNA (794 bp ND4). The red rectangles indicate the branches that are well supported (i.e. posterior probability values over 95, and/or bootstrap values over 70; the precise values are provided in Fig. S3). Seven genetic lineages were determined: SA Inland (blue), SA Coast (pink), Western Victoria (purple), Central Victoria (green), Eastern Victoria (orange), Tasmania (yellow) and Australian Captial Territory (red). Photos by Jules Farquhar.
| ACT | EVIC | CVIC | WVIC | Tas. | SAInland | SACoast | ||
|---|---|---|---|---|---|---|---|---|
| ACT | – | 0.006 | 0.006 | 0.006 | 0.007 | 0.006 | 0.007 | |
| EVIC | 0.031 | – | 0.003 | 0.008 | 0.005 | 0.007 | 0.008 | |
| CVIC | 0.031 | 0.011 | – | 0.007 | 0.005 | 0.007 | 0.008 | |
| WVIC | 0.037 | 0.042 | 0.041 | – | 0.008 | 0.004 | 0.005 | |
| Tas. | 0.035 | 0.020 | 0.022 | 0.045 | – | 0.008 | 0.008 | |
| SAInland | 0.037 | 0.039 | 0.038 | 0.009 | 0.043 | – | 0.006 | |
| SACoast | 0.046 | 0.051 | 0.051 | 0.021 | 0.051 | 0.026 | – |
Locations are Australian Capital Territory (ACT), Eastern Victoria (EVIC), Central Victoria (CVIC), Western Victoria (WVIC), Tasmania (Tas.), inland South Australia (SAInland), coastal South Australia (SACoast).
Discussion
Our results indicate that the glossy grass skink reaches sexual maturity in approximately 3 years, at a body size of around 40 mm SVL, and has a generation length of 5 years. Our estimate for the size at sexual maturity is consistent with that suggested by Hutchinson and Donnellan (1988) when the species was formally described. Indeed, the life history traits for the glossy grass skink are similar to that reported for other similar-sized viviparous skink species in south-eastern Australia (Hutchinson and Donnellan 1992; Chapple 2006; Atkins et al. 2020; Greer 2022). Importantly, our study represents the first detailed determination of the generation length of a Pseudemoia species (Greer 2022). Unfortunately, due to a limited number of specimens from alpine regions, we were unable to examine whether the life history traits of the glossy grass skink differed between the lowland (majority of the species range) and highland (alpine regions of Victoria, New South Wales, and the Australian Capital Territory) populations of the species. However, this possibility warrants further investigation as previous studies of viviparous skinks species in south-eastern Australia have documented substantial differences in life-history between high and low elevation populations (Rohr 1997; Wapstra et al. 2001; Atkins et al. 2020; Van Dyke et al. 2021).
We found that sexual dimorphism was evident in the glossy grass skink, with females having larger bodies and interlimb lengths, and males exhibiting larger heads and longer legs. Within squamates, it has been suggested that there is a strong selective pressure for females to have larger body size, and specifically longer interlimb lengths, in order to accommodate more developing eggs and/or young (Cox et al. 2003; Scharf and Meiri 2013; Meiri et al. 2020). Indeed, this pattern appears to hold in the glossy grass skink as litter size was positively correlated to female body size. Although the mean litter size for the glossy grass skink found in our study was slightly lower (4.6 vs 5.6) than that reported previously for the species by Hutchinson and Donnellan (1988), we found the species to have a broader range of litter sizes than previously reported (1–9 vs 4–9). The larger head size of male glossy grass skinks is concordant with previous reports in other squamates (Cox et al. 2003; Scharf and Meiri 2013), including viviparous skinks in south-eastern Australia (Clemann et al. 2004; Chapple 2006). Previous hypotheses for the selective advantage of males having larger heads have focused on the ability to grip females during copulation, or the importance of head size in determining the outcome of male–male competition and fights (Vitt and Cooper 1985; Cox et al. 2003; Scharf and Meiri 2013). Although it has previously been reported that male glossy grass skinks have longer limbs than females (Hutchinson and Donnellan 1992), the significance of this morphological dimorphism is unknown and warrants further investigation.
Our study provides information that will assist in assessing the glossy grass skink against IUCN Red List criteria. As species respond to changes in their environment at different rates, an estimate of generation length is an important component of most Red List criteria (Mace et al. 2008; Bird et al. 2020). Generation length, which incorporates information on the age at sexual maturity and lifespan, is important when considering population trends, and determining the capacity of the species to withstand threats or recover from population declines (Isaac et al. 2005; Rowe 2008; Bird et al. 2020). In addition, knowledge of litter size can provide information on the potential population growth of the species, whilst taking into account the potential for lower fecundity in younger (i.e. smaller) adults (Bird et al. 2020; Meiri et al. 2020). In regard to the glossy grass skink, it has a longer generation time than many smaller size skinks in south-eastern Australia (Greer 2022), and therefore might take longer to recover from population declines, making it more susceptible to a range of threats (e.g. Senior et al. 2021). However, its generation length, and thus intrinsic susceptibility to threats, may be consistent with most other similar-sized viviparous skinks in south-eastern Australia (Wapstra et al. 2001; Chapple 2006; Atkins et al. 2020).
Our study, combined with the results from Haines et al. (2014), indicates that the glossy grass skink is a single widespread, but genetically variable, species. This is because the level of genetic divergence observed in the species was less than, or equivalent to, the level of intraspecific divergence reported for a range of other Australian skink species (Chapple et al. 2004, 2005, 2011a, 2011b; Senior et al. 2022). Although some degree of morphological variation was evident within the glossy grass skink, there were no consistent morphological differences among the seven genetic lineages in the species. Interestingly, the location of the seven genetic lineages within the glossy grass skink were largely concordant with the recognised biogeographic boundaries in south-eastern Australia (Chapple et al. 2005, 2011a, 2011b; Bryant and Krosch 2016; Dissanayake et al. 2022). Assuming 2% sequence divergence per million years, as used in other Australian skinks (Chapple et al. 2005; Haines et al. 2014), this would indicate a divergence time of between 0.5 million years ago and 2.6 million years, ago during the Pliocene–Pleistocene boundary (Chapple et al. 2005; Dissanayake et al. 2022; Senior et al. 2022). This period was characterised by climatic fluctuations, and associated fluctuations in sea level (Byrne et al. 2011; Chapple et al. 2011a; Haines et al. 2014). The glossy grass skink, and other Pseudemoia species, tend to inhabit areas with high precipitation (Hutchinson and Donnellan 1992; Haines et al. 2014), and consequently drying due to climatic oscillations may present geographic boundaries to populations (Byrne et al. 2011; Chapple et al. 2011a; Bryant and Krosch 2016). In particular, an east–west split in Victoria is common in many skink species, and is likely due to historical volcanic activity (Chapple et al. 2005; Senior et al. 2022). Additionally, alpine populations are often separate from nearby lowland populations due largely to climatic and resource variation (Chapple et al. 2005, 2011a). Overall, our genetic data might indicate that the glossy grass skink may not disperse across fragmented landscapes, and therefore habitat loss and fragmentation may result in decreased connectivity and genetic diversity within the species (e.g. Crain et al. 2008; Dixo et al. 2009; Haines et al. 2014; Senior et al. 2022). However, further work involving more detailed sampling within populations and across the species range, along with additional nuclear loci, would be required to confirm this result.
Conclusions
We have provided a case study for how a targeted, multifaceted research project can be effective at rapidly gathering data that can be used to contribute vital information to the assessment of extinction risk in Data Deficient species. This approach can be broadly applicable to any Data Deficient species, where there are sufficient museum specimens, and tissue samples, available. For the glossy grass skink, we have determined the generation length of the species, providing information that will allow the species to be assessed against Criteria A, C and E on the IUCN Red List (IUCN Standards and Petitions Committee 2022). Our molecular data, combined with recent field-based studies on the species (Farquhar et al. 2024), indicates that the species may be susceptible to habitat loss and fragmentation (Criterion B), and provides information (i.e. several distinct genetic lineages) that may be useful for determining the number of subpopulations that are present in the species (Criteria B and C). In a recent related study, we clarified the distribution of the glossy grass skink, used field surveys to determine that the species has disappeared at ~50% of historical sites, and showed that a third of the species’ predicted range occurs in cleared agricultural land that is now likely unsuitable for the species (Farquhar et al. 2023) (Criteria A, B and C). These findings, combined with the results of the current study, will allow the species to be assessed against IUCN Red List criteria, and will most likely result in the species being listed as Vulnerable under Criterion B.
Data availability
The data and code associated with this study are provided in the online supplementary material. Our DNA sequence data has been submitted to GenBank under the accession numbers provided in Table 1.
Declaration of funding
This work was supported by a grant from the Australian Research Council (FT200100108; to DGC).
Author contributions
Lucy Wotherspoon: Conceptualisation, Methodology, Formal analysis, Investigation, Data curation, Writing – Original Draft, Visualisation; Maggie Haines: Conceptualisation, Methodology, Formal analysis, Investigation, Data curation, Writing – Review and Editing, Visualisation, Supervision; Jules Farquhar: Conceptualisation, Methodology, Formal analysis, Investigation, Data curation, Writing – Review and Editing, Visualisation, Supervision; David Chapple: Conceptualisation, Methodology, Formal analysis, Investigation, Data curation, Writing – Original Draft, Visualisation, Supervision, Project administration, Funding acquisition.
Acknowledgements
We thank Jo Sumner and Museums Victoria, and Mark Hutchinson and the South Australia Museum for providing preserved specimens and tissues, and their facilities. We thank Luke Bonifacio for providing laboratory assistance.
References
Atkins ZS, Clemann N, Chapple DG, Edwards AM, Sinsch U, Hantzschmann AM, Schroder M, Scroggie MP, Robert KA (2020) Demographic and life history variation in two sky-island populations of an endangered alpine lizard. Journal of Zoology 310, 34-44.
| Crossref | Google Scholar |
Barnosky AD, Matzke N, Tomiya S, Wogan GOU, Swartz B, Quental TB, Marshall C, McGuire JL, Lindsey EL, Maguire KC, Mersey B, Ferrer EA (2011) Has the earth’s sixth mass extinction already arrived? Nature 471, 51-57.
| Crossref | Google Scholar | PubMed |
Barter M, Bonifacio LR, Duran A, Goulet CT, Tingley R, Shea GM, Meiri S, Chapple DG (2022) Predictors of geographic range size in Australian skinks. Global Ecology and Biogeography 31, 113-122.
| Crossref | Google Scholar |
Bird JP, Martin R, Akçakaya HR, Gilroy J, Burfield IJ, Garnett ST, Symes A, Taylor J, Şekercioğlu ÇH, Butchart SHM (2020) Generation lengths of the world’s birds and their implications for extinction risk. Conservation Biology 34, 1252-1261.
| Crossref | Google Scholar | PubMed |
Bland LM, Böhm M (2016) Overcoming data deficiency in reptiles. Biological Conservation 204, 16-22.
| Crossref | Google Scholar |
Bland LM, Bielby J, Kearney S, Orme CDL, Watson JEM, Collen B (2017) Toward reassessing data-deficient species. Conservation Biology 31, 531-539.
| Crossref | Google Scholar | PubMed |
Bryant LM, Krosch MN (2016) Lines in the land: a review of evidence for eastern Australia’s major biogeographical barriers to closed forest taxa. Biological Journal of the Linnean Society 119, 238-264.
| Crossref | Google Scholar |
Byrne M, Steane DA, Joseph L, Yeates DK, Jordan GJ, Crayn D, et al. (2011) Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. Journal of Biogeography 38, 1635-1656.
| Crossref | Google Scholar |
Caetano GHdO, Chapple DG, Grenyer R, Raz T, Rosenblatt J, Tingley R, Böhm M, Meiri S, Roll U (2022) Automated assessment reveals that the extinction risk of reptiles is widely underestimated across space and phylogeny. PLoS Biology 20, e3001544.
| Crossref | Google Scholar |
Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM (2015) Accelerated modern human-induced species losses: entering the sixth mass extinction. Science Advances 1, e1400253.
| Crossref | Google Scholar | PubMed |
Ceballos G, Ehrlich PR, Raven PH (2020) Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. Proceedings of the National Academy of Sciences of the United States of America 117, 13596-13602.
| Crossref | Google Scholar | PubMed |
Chan KO, Grismer LL (2022) GroupStruct: an R package for allometric size correction. Zootaxa 5124, 471-482.
| Crossref | Google Scholar | PubMed |
Chapple DG (2006) Life history and reproductive ecology of White’s skink, Egernia whitii. Australian Journal of Zoology 53, 353-360.
| Crossref | Google Scholar |
Chapple DG, Keogh JS, Hutchinson MN (2004) Molecular phylogeography and systematics of the arid-zone members of the Egernia whitii (Lacertilia: Scincidae) species group. Molecular Phylogenetics and Evolution 33, 549-561.
| Crossref | Google Scholar | PubMed |
Chapple DG, Keogh JS, Hutchinson MN (2005) Substantial genetic substructuring in southeastern and alpine Australia revealed by molecular phylogeography of the Egernia whitii (Lacertilia: Scincidae) species group. Molecular Ecology 14, 1279-1292.
| Crossref | Google Scholar | PubMed |
Chapple DG, Ritchie PA, Daugherty CH (2009) Origin, diversification, and systematics of the New Zealand skink fauna (Reptilia: Scincidae). Molecular Phylogenetics and Evolution 52, 470-487.
| Crossref | Google Scholar | PubMed |
Chapple DG, Hoskin CJ, Chapple SNJ, Thompson MB (2011a) Phylogeographic divergence in the widespread delicate skink (Lampropholis delicata) corresponds to dry habitat barriers in eastern Australia. BMC Evolutionary Biology 11, 191.
| Crossref | Google Scholar | PubMed |
Chapple DG, Chapple SNJ, Thompson MB (2011b) Biogeographic barriers in south-eastern Australia drive phylogeographic divergence in the garden skink, Lampropholis guichenoti. Journal of Biogeography 38, 1761-1775.
| Crossref | Google Scholar |
Clemann N, Chapple DG, Wainer J (2004) Sexual dimorphism, diet, and reproduction in the swamp skink, Egernia coventryi. Journal of Herpetology 38, 461-467.
| Crossref | Google Scholar |
Cox RM, Skelly SL, John-Alder HB (2003) A comparative test of adaptive hypotheses for sexual size dimorphism in lizards. Evolution 57, 1653-1669 10.1111/j.0014-3820.2003.tb00371.x.
| Google Scholar | PubMed |
Crain CM, Kroeker K, Halpern BS (2008) Interactive and cumulative effects of multiple human stressors in marine systems. Ecology Letters 11, 1304-1315.
| Crossref | Google Scholar | PubMed |
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9, 772.
| Crossref | Google Scholar | PubMed |
Dissanayake DSB, Holleley CE, Sumner J, Melville J, Georges A (2022) Lineage diversity within a widespread endemic Australian skink to better inform conservation in response to regional-scale disturbance. Ecology and Evolution 12, e8627.
| Crossref | Google Scholar | PubMed |
Dixo M, Metzger JP, Morgante JS, Zamudio KR (2009) Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic Coastal Forest. Biological Conservation 142, 1560-1569.
| Crossref | Google Scholar |
Doherty TS, Glen AS, Nimmo DG, Ritchie EG, Dickman CR (2016) Invasive predators and global biodiversity loss. Proceedings of the National Academy of Sciences 113, 11261-11265.
| Crossref | Google Scholar |
Du W, Ji X, Shine R (2005) Does body-volume constrain reproductive output in lizards? Biology Letters 1, 98-100.
| Crossref | Google Scholar | PubMed |
Farquhar JE, Carlesso A, Pili A, Gale N, Chapple DG (2023) Capturing uncatalogued distribution records to improve conservation assessments of data-deficient species: a case study using the glossy grass skink. Animal Conservation 27, 124-137.
| Crossref | Google Scholar |
Farquhar JE, Wotherspoon L, Porter H, Chapple DG (2024) Habitat loss and degradation reduce abundance of the glossy grass skink, Pseudemoia rawlinsoni. Wildlife Research
| Google Scholar |
Fung HC, Waples RS (2017) Performance of IUCN proxies for generation length. Conservation Biology 31, 883-893.
| Crossref | Google Scholar | PubMed |
Gillespie G, Clemann N, Hutchinson M, Michael D, Melville J, Robertson P, Chapple DG (2018) Pseudemoia rawlinsoni, The IUCN Red List of Threatened Species 2018: e.T109480985A109480994. Available at https://www.iucnredlist.org/species/109480985/109480994 [Accessed 15 October 2022]
Graham KA, Mahony SV, Chapple DG, Farquhar JE (2023) The long unknown: rediscovery of the long sunskink, Lampropholis elongata (Squamata: Scincidae), after almost a decade, and after 50 years of data deficiency. Austral Ecology 48, 877-884.
| Crossref | Google Scholar |
Greer AE (1982) A new species of Leiolopisma (Lacertilia: Scincidae) from Western Australia. Records of the Australian Museum 34, 549-573.
| Crossref | Google Scholar |
Greer AE (2022) Encyclopedia of Australian reptiles. Version 1 October 2022. Available at https://www.researchgate.net/publication/364060690_Encyclopedia_of_Australian_Reptiles_Version_1_October_2022 [Accessed 15 October 2022]
Gumbs R, Gray CL, Böhm M, Hoffmann M, Grenyer R, Jetz W, et al. (2020) Global priorities for conservation of reptilian phylogenetic diversity in the face of human impacts. Nature Communications 11, 2616.
| Crossref | Google Scholar | PubMed |
Haines ML, Moussalli A, Stuart-Fox D, Clemann N, Melville J (2014) Phylogenetic evidence of historic mitochondrial introgression and cryptic diversity in the genus Pseudemoia (Squamata: Scincidae). Molecular Phylogenetics and Evolution 81, 86-95.
| Crossref | Google Scholar | PubMed |
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95-98.
| Google Scholar |
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754-755.
| Crossref | Google Scholar | PubMed |
Hutchinson MW, Donnellan SC (1988) A new species of scincid lizard related to Leiolopisma entrecasteauxii, from Southeastern Australia. Royal Society of South Australia 112, 143-151.
| Google Scholar |
Hutchinson MN, Donnellan SC (1992) Taxonomy and genetic variation in the Australian lizards of the genus Pseudemoia (Scincidae: Lygosominae). Journal of Natural History 26, 215-264.
| Crossref | Google Scholar |
IPBES (2019) Global assessment report on biodiversity and ecosystem services of the intergovernmental science-policy platform on biodiversity and ecosystem services. (Eds ES Brondizio, J Settele, S Díaz, HT Ngo) p. 1148. Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services, IPBES Secretariat, Bonn, Germany. doi:10.5281/zenodo.3831673
Isaac NJB, Jones KE, Gittleman JL, Purvis A (2005) Correlates of species richness in mammals: body size, life history, and ecology. The American Naturalist 165, 600-607.
| Crossref | Google Scholar | PubMed |
IUCN Standards and Petitions Committee (2022) Guidelines for using the IUCN red list categories and criteria. Version 15.1. Prepared by the Standards and Petitions Committee. Available at https://www.iucnredlist.org/documents/RedListGuidelines.pdf
Johnson CN, Balmford A, Brook BW, Buettel JC, Galetti M, Guangchun L, Wilmshurst JM (2017) Biodiversity losses and conservation responses in the Anthropocene. Science 356, 270-275.
| Crossref | Google Scholar | PubMed |
Laver RJ, Purwandana D, Ariefiandy A, Imansyah J, Forsyth D, Ciofi C, Jessop TS (2012) Life-history and spatial determinants of somatic growth dynamics in Komodo dragon populations. PLoS ONE 7, e45398.
| Crossref | Google Scholar | PubMed |
Maayan I, Reynolds RG, Goodman RM, Hime PM, Bickel R, Luck EA, Losos JB (2022) Fixation and preservation contribute to distortion in vertebrate museum specimens: a 10-year study with the lizard Anolis sagrei. Biological Journal of the Linnean Society 136, 443-454.
| Crossref | Google Scholar |
Mace GM, Collar NJ, Gaston KJ, Hilton-Taylor C, Akçakaya HR, Leader-Williams N, Milner-Gulland EJ, Stuart SN (2008) Quantification of extinction risk: IUCN’s system for classifying threatened species. Conservation Biology 22, 1424-1442.
| Crossref | Google Scholar | PubMed |
Meiri S, Avila L, Bauer AM, Chapple DG, Das I, Doan TM, Doughty P, Ellis R, Grismer L, Kraus F, Morando M, Oliver P, Pincheira-Donoso D, Ribeiro-Junior MA, Shea G, Torres-Carvajal O, Slavenko A, Roll U (2020) The global diversity and distribution of lizard clutch sizes. Global Ecology and Biogeography 29, 1515-1530.
| Crossref | Google Scholar |
Olsson M, Shine R (1996) Does reproductive success increase with age or with size in species with indeterminate growth? A case study using sand lizards (Lacerta agilis). Oecologia 105, 175-178.
| Crossref | Google Scholar | PubMed |
Patnaik BK (1994) Ageing in reptiles. Gerontology 40, 200-220.
| Crossref | Google Scholar | PubMed |
Pecl GT, Araújo MB, Bell JD, Blanchard J, Bonebrake TC, Chen I-C, et al. (2017) Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 355, eaai9214 10.1126/science.aai921.
| Google Scholar |
Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO (2014) The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752.
| Crossref | Google Scholar |
R Core Team (2021) ‘R: a language and environment for statistical computing.’ (R Foundation for Statistical Computing: Vienna, Austria) Available at https://www.R-project.org/ [Accessed 15 October 2022]
Rohr DH (1997) Demographic and life-history variation in two proximate populations of a viviparous skink separated by a steep altitudinal gradient. The Journal of Animal Ecology 66, 567-578.
| Crossref | Google Scholar |
Rowe CL (2008) “The calamity of so long life”: life histories, contaminants, and potential emerging threats to long-lived vertebrates. BioScience 58, 623-631.
| Crossref | Google Scholar |
Scharf I, Meiri S (2013) Sexual dimorphism of heads and abdomens: different approaches to ‘being large’ in female and male lizards. Biological Journal of the Linnean Society 110, 665-673.
| Crossref | Google Scholar |
Senior AF, Böhm M, Johnstone CP, McGee MD, Meiri S, Chapple DG, Tingley R (2021) Correlates of extinction risk in Australian squamate reptiles. Journal of Biogeography 48, 2144-2152.
| Crossref | Google Scholar |
Senior AF, Clemann N, Gardner MG, Harrisson KA, While GM, Chapple DG (2022) Genetic structure, diversity and distribution of a threatened lizard affected by widespread habitat fragmentation. Conservation Genetics 23, 151-165.
| Crossref | Google Scholar |
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-1313.
| Crossref | Google Scholar | PubMed |
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of the United States of America 101, 11030-11035.
| Crossref | Google Scholar |
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38, 3022-3027.
| Crossref | Google Scholar | PubMed |
Taylor EH (1935) A taxonomic study of the cosmopolitan scincoid lizards of the genus Eumeces with an account of the distribution and relationships of its species. Bulletin of The University of Kansas 36, 70-80.
| Google Scholar |
Van Dyke JU, Thompson MB, Burridge CP, Castelli MA, Clulow S, Dissanayake DSB, Dong CM, Doody JS, Edwards DL, Ezaz T, Friesen CR, Gardner MG, Georges A, Higgie M, Hill PL, Holleley CE, Hoops D, Hoskin CJ, Merry DL, Riley JL, Wapstra E, While GM, Whiteley SL, Whiting MJ, Zozaya SM, Whittington CM (2021) Australian lizards are outstanding models for reproductive biology research. Australian Journal of Zoology 68, 168-199.
| Crossref | Google Scholar |
Vervust B, Van Dongen S, Van Damme R (2009) The effect of preservation on lizard morphometrics – an experimental study. Amphibia-Reptilia 30, 321-329.
| Crossref | Google Scholar |
Vitt LJ, Cooper WE, Jr (1985) The evolution of sexual dimorphism in the skink Eumeces laticeps: an example of sexual selection. Canadian Journal of Zoology 63, 995-1002.
| Crossref | Google Scholar |
Wapstra E, Swain R, O’Reilly JM (2001) Geographic variation in age and size at maturity in a small Australian viviparous skink. Copeia 2001, 646-655.
| Crossref | Google Scholar |
Wotherspoon L, de Oliveira Caetano GH, Roll U, Meiri S, Pili A, Tingley R, Chapple DG (2024) Inferring the extinction risk of data deficient and not evaluated Australian squamates. Austral Ecology 49, e13485.
| Crossref | Google Scholar |


