Evidence of long-distance movement of green sawfish (Pristis zijsron) in Western Australia
Jack Ingelbrecht A , Mark G. Allen A , Rebecca L. Bateman A , Brendan C. Ebner A B , Travis Fazeldean A , Kurt N. Krispyn A , Karissa O. Lear
A , Mark G. Allen A , Rebecca L. Bateman A , Brendan C. Ebner A B , Travis Fazeldean A , Kurt N. Krispyn A , Karissa O. Lear  A , Tegan Lee A , Alan J. Lymbery A , Rory B. McAuley C , Nicole M. Phillips D , Jeff M. Whitty A , Barbara E. Wueringer E F and David L. Morgan A *
A , Tegan Lee A , Alan J. Lymbery A , Rory B. McAuley C , Nicole M. Phillips D , Jeff M. Whitty A , Barbara E. Wueringer E F and David L. Morgan A *
A
B
C
D
E
F
Abstract
In this study, single-nucleotide polymorphisms (SNPs) were used to investigate kinship for the green sawfish (Pristis zijsron).
To examine the relatedness of P. zijsron across an expansive coastline in Western Australia.
Sampling was conducted between the Fitzroy River estuary and Bay of Rest in the eastern Indian Ocean (north-western Australia) between 2003 and 2022. SNPs were generated from tissues collected from 137 live and 1 recently deceased P. zijsron.
Overall, 62 individual P. zijsron were assigned to 25 litters of full siblings, with litter sizes ranging from 2 to 5 pups, and 76 P. zijsron individuals were assigned to 96 half sibling pairwise relationships. Four pairs of half siblings were captured more than 500 km and born at least 6 years apart, including one pair of neonates captured ~870 km and 8 years apart, in the Ashburton River estuary (Pilbara) and Cable Beach (Broome). Furthermore, a pair of full-sibling pups (i.e. young of the year) caught at Cape Keraudren (Pilbara) in 2008 were half siblings of a pup caught in the Ashburton River in 2014.
This study provides evidence of long-distance, likely parental, movement of P. zijsron.
Dispersal of P. zijsron over large spatial scales indicates that populations could be replenished from elsewhere should they experience a decline, thereby reducing the risk of localised extinction for this species.
Keywords: dispersal, Kimberley, Pedigree, Pilbara, Pristidae, reproduction, Rhinopristiformes, sibship, single-nucleotide polymorphism.
Introduction
In conservation biology, kinship information is useful for identifying patterns of movement, reproductive behaviour, and estimating inbreeding, which can help establish the extinction risk of threatened populations (Madsen et al. 1996; Goudet et al. 2018). However, pedigrees (i.e. records of descent) for wild populations are often difficult, if not impossible, to obtain from direct observations alone, particularly for extinction-prone species that typically have low abundances and restricted ranges (Mace et al. 2008; Jones and Wang 2010a). Fortunately, because of recent advances in next-generation sequencing technology and computational power, the ability to statistically infer pedigrees from genomic data has greatly improved over the past decade (Jones and Wang 2010a, 2010b; Huisman 2017; Goudet et al. 2018).
The sawfishes (Pristidae) are a group of large-bodied, charismatic elasmobranchs, which are among the most threatened marine fishes globally (Dulvy et al. 2016). All five extant species are designated as Critically Endangered on the International Union for Conservation of Nature’s Red List of Threatened Species (Carlson et al. 2022; Espinoza et al. 2022; Grant et al. 2022; Harry et al. 2022; Haque et al. 2023), having experienced a 30–81% decline in extent of occurrence over the past century, primarily because of overexploitation by fisheries (Dulvy et al. 2016). The green sawfish (Pristis zijsron Bleeker, 1851) is one of the largest fishes, attaining lengths of at least 5.4 m in Australian waters (Peverell 2005; Morgan et al. 2011) and possibly in excess of 7 m overall (Compagno and Last 1998; Last and Stevens 2009). Viable populations of this species are mostly restricted to northern Australia (Peverell 2005; Morgan et al. 2011), with previous kinship investigations for P. zijsron having been limited to individuals in the Ashburton River estuary and adjacent tidal creeks, Western Australia (see Ingelbrecht et al. 2024). North-western Australia is a globally important refuge for P. zijsron, with the Ashburton River estuary and adjacent tidal creeks in particular representing one of the world’s most important nursery areas for this species (Morgan et al. 2011, 2015, 2017). In this study, single-nucleotide polymorphisms (SNPs) were used to infer kinship among P. zijsron captured between the Fitzroy River and the Bay of Rest, Western Australia, to better understand the ecological connectivity of this species and establish more effective conservation strategies along the remote north-western coastline of Australia.
Materials and methods
Ethics approval
Handling and sampling of sawfish was conducted under Murdoch University Animal Ethics Approvals RW2397/11 and RW3191/19, Western Australian Government Department of Primary Industries and Regional Development Fisheries Exemption numbers 3378 and 3553, Department of Fisheries Regulation 178 (SPA 11-11), and Department of Environment and Conservation Permit SF007889.
Sample collection
Pristis zijsron tissue samples were collected from individuals at eight sites in Western Australia (Fig. 1, Table 1). Live P. zijsron were captured with monofilament gillnets (see Morgan et al. 2015) or cast nets. Upon capture, P. zijsron were placed onto their backs in the shallows with their gills submerged, inducing a state of tonic immobility, and then measured for stretched total length (STL) and sexed. Tissue samples in the form of fin clips or hole punches were taken from either the pelvic or dorsal fins and immediately preserved in 100% ethanol. Captured sawfish were then released at the site of capture. Samples were collected from 137 live juvenile P. zijsron (Table 1) between 2003 and 2022 (651–3195 mm STL), and 1 carcass from Cable Beach in 2022, measured at 875 mm STL, which is consistent with the reported size at birth for this species (i.e. 750–900 mm) (see Morgan et al. 2015; Lear et al. 2023). Most of these were from the Ashburton River estuary and adjacent tidal creeks, which had been included in previous kinship investigations (see Ingelbrecht et al. 2024), with the present study focusing instead on individual relationships between different populations of P. zijsron along the north-western Australian coastline.
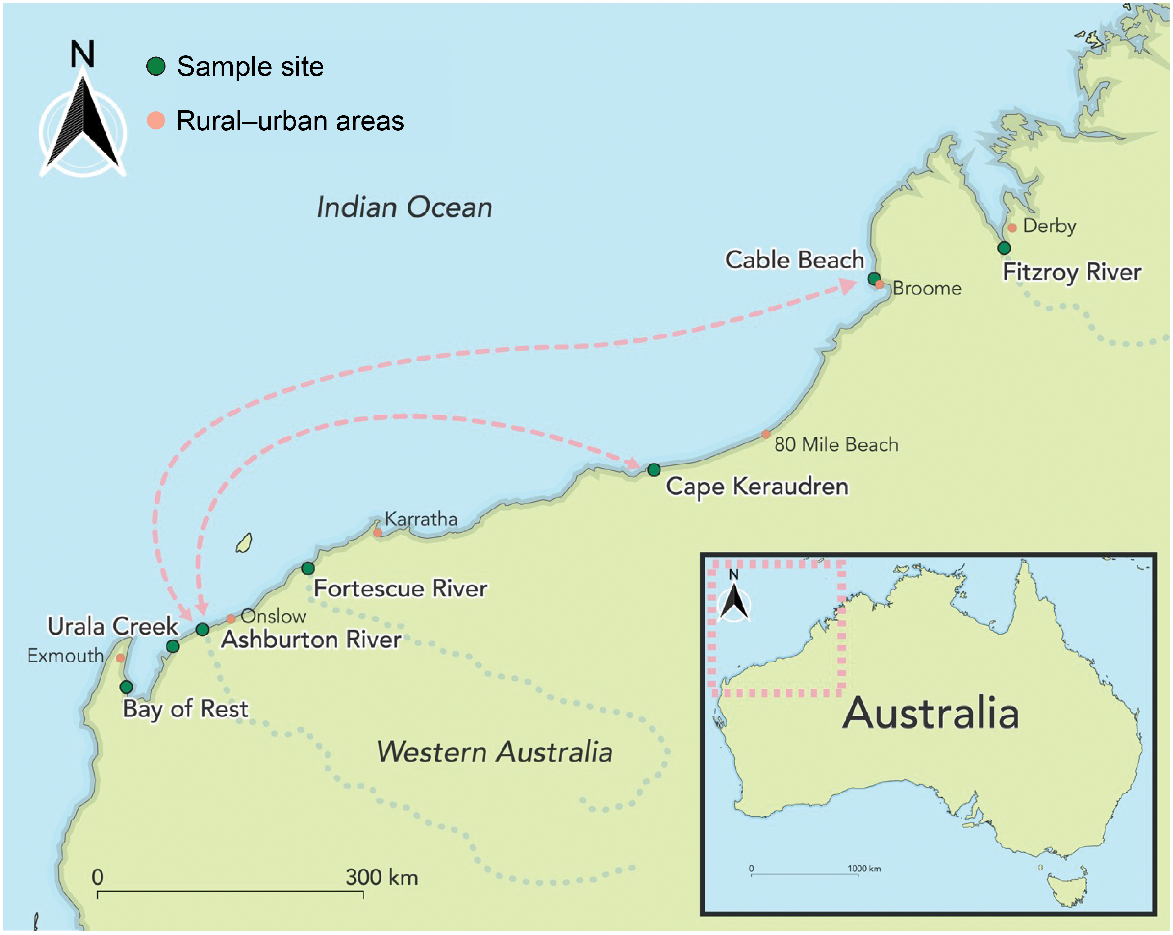
Sampling locations for green sawfish (Pristis zijsron) in Western Australia, illustrating the distance between capture locations of half siblings (arrows) captured at Cape Keraudren in 2008 and the Ashburton River in 2014, or the Ashburton River in 2014 and Cable Beach in 2022.
| Site | n | Sex | STL (mm) | Age class | Capture months and years | |||
|---|---|---|---|---|---|---|---|---|
| F | M | 0+ | ≥1 | |||||
| Fitzroy River | 1 | 1 | – | 1567 | – | 1 | September 2015 | |
| Cable Beach | 1 | – | 1 | 875 | 1 | – | October 2022 | |
| 80 Mile Beach | 4 | – | 4 | 1120–2090 | 3 | 1 | April 2003–June 2004 | |
| Cape Keraudren | 12 | 3 | 9 | 1000–2600 | 4 | 8 | April 2008 | |
| Fortescue River | 2 | – | 2 | 2001–2015 | – | 2 | August 2022 | |
| Ashburton River | 114 | 61 | 53 | 651–3195 | 77 | 37 | April 2011–November 2022 | |
| Urala Creek | 1 | – | 1 | 1225 | 1 | – | February 2019 | |
| Bay of Rest | 3 | – | 3 | 804–843 | 3 | – | September 2021 | |
Ashburton River includes samples from adjacent Ashburton Delta, Four Mile Creek and Hooley Creek. Age classes are based on Lear et al. (2023). F, female; M, male; STL, stretched total length; 0+, young of the year; ≥1, juveniles at least 1-year old.
Genotyping and data filtering
Tissue samples were sectioned into ~10-mg pieces and submerged in ~100 μL of 70% ethanol in an Eppendorf Twintec polymerase chain reaction (PCR) extraction plate, in preparation for DNA extraction and genotyping at Diversity Arrays Technology (DArT, DArT Pty Ltd, Canberra, ACT, Australia). Extraction of DNA was performed with a NucleoMag magnetic bead-based extraction kit (Macherey–Nagel, Berlin, Germany), by using the methods described by Ingelbrecht et al. (2024). Genotyping was performed using Pristis DArTseq (DArT Pty Ltd), with a high-density assay and sequencing effort of 2.5 million reads. Single-read sequencing was performed on a Novaseq X high-throughput sequencer (Illumina, San Diego, CA, USA), following standard protocols described by Kilian et al. (2012). To assess the quality of markers and consistency of allele calling (i.e. reproducibility), 30% of sampled individuals were genotyped in technical replication.
Data for 130 individuals, corresponding with 35,210 SNPs, were generated by DArT Pty Ltd. Samples from eight P. zijsron failed sequencing and were therefore not included. Data filtering was implemented in R (ver. 4.2.3, R Foundation for Statistical Computing, Vienna, Austria, see https://www.r-project.org/) using the package dartR (ver. 2.7.2, see https://cran.r-project.org/package=dartR; Gruber et al. 2018). For statistical inference of kinship, the use of at least 500 SNPs with a moderately high minor allele frequency (MAF) (i.e. ≥0.2) is recommended for maximising pedigree assignment rate, while minimising the rate of errors (Huisman 2017; Premachandra et al. 2019). Therefore, SNPs were retained according to call rate (≥95%), reproducibility (i.e. the proportion of identical genotypes calculated from technical replicates; ≥ 95%), and read depth (lower/upper thresholds = 5/40). Because allele frequency affects the accuracy and rate of kinship inference (see Huisman 2017; Premachandra et al. 2019), four different MAF lower thresholds were used (≥0.1, 0.2, 0.3 or 0.4). Furthermore, because of the impacts of MAF filtering on heterozygosity, different individual sample heterozygosity lower/upper thresholds were applied depending on MAF, as follows: 0/0.42 (MAF ≥ 0.1), 0/0.47 (MAF ≥ 0.2), 0/0.52 (MAF ≥ 0.3), 0/0.55 (MAF ≥ 0.4), generating four filtered SNP datasets. Monomorphic loci or loci showing significant departure from Hardy–Weinberg equilibrium (HWE) (at a significance threshold of α = 0.05, using a Bonferroni correction for multiple comparisons) were removed.
Kinship analysis
Kinship was inferred using the following three independent analyses: Sequoia (ver. 2.5.3, see https://cran.r-project.org/package=sequoia; Huisman 2017), COLONY (ver. 2.0.6.8, see https://www.zsl.org/about-zsl/resources/software/colony; Jones and Wang 2010b), and genomic relatedness probabilities calculated in dartR. Kinship assignments through Sequoia and COLONY were performed by full-pedigree reconstructions, by using filtered datasets of SNPs and two genotyping error rates: 1 or 5%. Sequoia and COLONY pedigrees were reconstructed using the same parameters as described previously for P. zijsron (see Ingelbrecht et al. 2024), but using a threshold log-likelihood ratio (LLR) ‘Tfilter’ of −4 in Sequoia.
Genomic relatedness probabilities, calculated from all available SNPs, were used to corroborate kinship assignments and distinguish between first- and second-degree relatives. Pairwise genomic relatedness probabilities between 0.41–0.59 and 0.18–0.31 were taken to indicate first- and second-degree relationships respectively, on the basis of 95% confidence intervals for relatedness reported by Speed and Balding (2015). The most likely genealogical relationships were assembled manually, using combined information from Sequoia and COLONY pedigrees, and estimates of pairwise genomic relatedness. To account for false positive inferences, an assignment was accepted (see reported assignments) only if it was supported by both Sequoia and COLONY in at least one pedigree, or if these analyses were not in congruence, by one of these analyses and pairwise genomic relatedness probabilities. False-negative assignments were accounted for by reconstructing pedigrees from multiple SNP datasets with varying MAF values (≥0.1, 0.2, 0.3 or 0.4). Furthermore, an assignment was deemed to be supported by COLONY only if it was accompanied by a confidence probability of ≥90%. Acceptance of full-sibling assignments through COLONY was based on exclusive probabilities (i.e. the probability that all individuals within a family are full siblings and that no other individuals are full siblings with that family), whereas acceptance of half siblings was based on confidence probabilities of assigned dyads. For young-of-the-year (YOY) P. zijsron, capture location was assumed to be proximal to the location of birth. Age classes were estimated using growth rate information for P. zijsron in Western Australia (Lear et al. 2023). Full siblings estimated to be of the same age were assumed to be littermates.
Results and discussion
Filtering of genotype data for 130 P. zijsron resulted in the retention of 3459, 2399, 1439 or 673 SNPs when the MAF was ≥0.1, 0.2, 0.3 or 0.4 respectively. Two P. zijsron were discarded for having heterozygosities that exceeded the upper filter thresholds. Data for one additional P. zijsron were not included in the kinship analyses because this individual was scored for less than 20% of SNPs. Of the initial 35,210 SNPs, 3684 were discarded for significant departure from HWE.
Sibling pairs
Overall, 62 P. zijsron were assigned to 25 litters of full siblings, with litter size ranging from two to five pups. Furthermore, 76 P. zijsron were assigned to 96 half-sibling pairs (i.e. pairwise relationships). Two full-sibling litters and six half-sibling pairs involved at least one P. zijsron sampled outside of the Ashburton River estuary and adjacent tidal creeks (Table 2). Of these, one individual was a half sibling of two other P. zijsron, which in turn were full siblings. These three individuals were captured together as neonates (804–843 mm STL) in the Bay of Rest, Exmouth Gulf, and are most likely to be littermates. Furthermore, one full-sibling pair was captured at Cape Keraudren in 2008, as YOY (i.e. 0+) (1000 and 1070 mm STL), providing the first evidence of P. zijsron parturition in the Exmouth Gulf and Cape Keraudren (Table 2).
| ID | Site | Distance (km) | Age class | Sex | STL (mm) | Catch months and years | Confidence probability (%) | |
|---|---|---|---|---|---|---|---|---|
| Full siblings | ||||||||
| 2008 #01 | Cape Keraudren | 0 | 0+ | M | 1070 | April 2008 | 100 | |
| 2008 #06 | Cape Keraudren | 0+ | M | 1000 | April 2008 | |||
| 2021 #24 | Bay of Rest | 0 | 0+ | M | 809 | September 2021 | 100 | |
| 2021 #26 | Bay of Rest | 0+ | M | 804 | September 2021 | |||
| Half siblings | ||||||||
| 2008 #01 | Cape Keraudren | ~530 | 0+ | M | 1070 | April 2008 | 100 | |
| 2014 #02 A | Ashburton River | 0+ | M | 874 | November 2014 | |||
| 2008 #06 | Cape Keraudren | ~530 | 0+ | M | 1000 | April 2008 | 94.6 | |
| 2014 #02 A | Ashburton River | 0+ | M | 874 | November 2014 | |||
| 2008 #04 | Cape Keraudren | ~530 | 1+ | M | 1560 | April 2008 | 100 | |
| 2014 #05 | Ashburton River | 1+ | F | 1620 | November 2014 | |||
| 2014 #03 | Ashburton River | ~870 | 0+ | F | 846 | November 2014 | 100 | |
| 2022 #38 | Cable Beach | 0+ | M | 875 | October 2022 | |||
| 2021 #24 | Bay of Rest | 0 | 0+ | M | 809 | September 2021 | 100 | |
| 2021 #25 A | Bay of Rest | 0+ | M | 843 | September 2021 | |||
| 2021 #25 A | Bay of Rest | 0 | 0+ | M | 843 | September 2021 | 100 | |
| 2021 #26 | Bay of Rest | 0+ | M | 804 | September 2021 | |||
Siblings are paired together, with different pairs separated by a change of shading. Identification number (ID) for each individual is the year followed by the order (#) the individual was caught. Distance refers to the straight-line distance separating capture locations of siblings. Age classes are based on Lear et al. (2023). Confidence values for assignments by COLONY (probabilities not available for Sequoia pedigrees) refer to exclusive probabilities for full siblings and dyad assignment probabilities for half siblings. 0+, young-of-the-year; 1+, juveniles older than 1 year; F, female; M, male; STL, stretched total length.
Distribution of kin over long distances
Four half-sibling pairs involved sawfish captured over 500 km apart (Table 2). Of these, one pair consisted of an individual captured in the Ashburton River estuary in 2014, as a neonate (846 mm STL), and its half sibling that was sampled as a recently deceased neonate (875 mm STL), ~870 km to the north-east at Cable Beach, Broome, in 2022 (i.e. ~8 years separating their births) (Fig. 2). The remaining three pairs (involving five juvenile P. zijsron) each consisted of one individual captured at Cape Keraudren in 2008, and their half siblings that were captured ~530 km to the south-west in the Ashburton River estuary in 2014, including one full-sibling pair from Cape Keraudren that shared the same half sibling from the Ashburton River (Table 2). These five individuals were estimated to be 2 years old or younger at the time they were captured (≤1620 mm STL), with ~6–7 years separating half-sibling births (Lear et al. 2023).
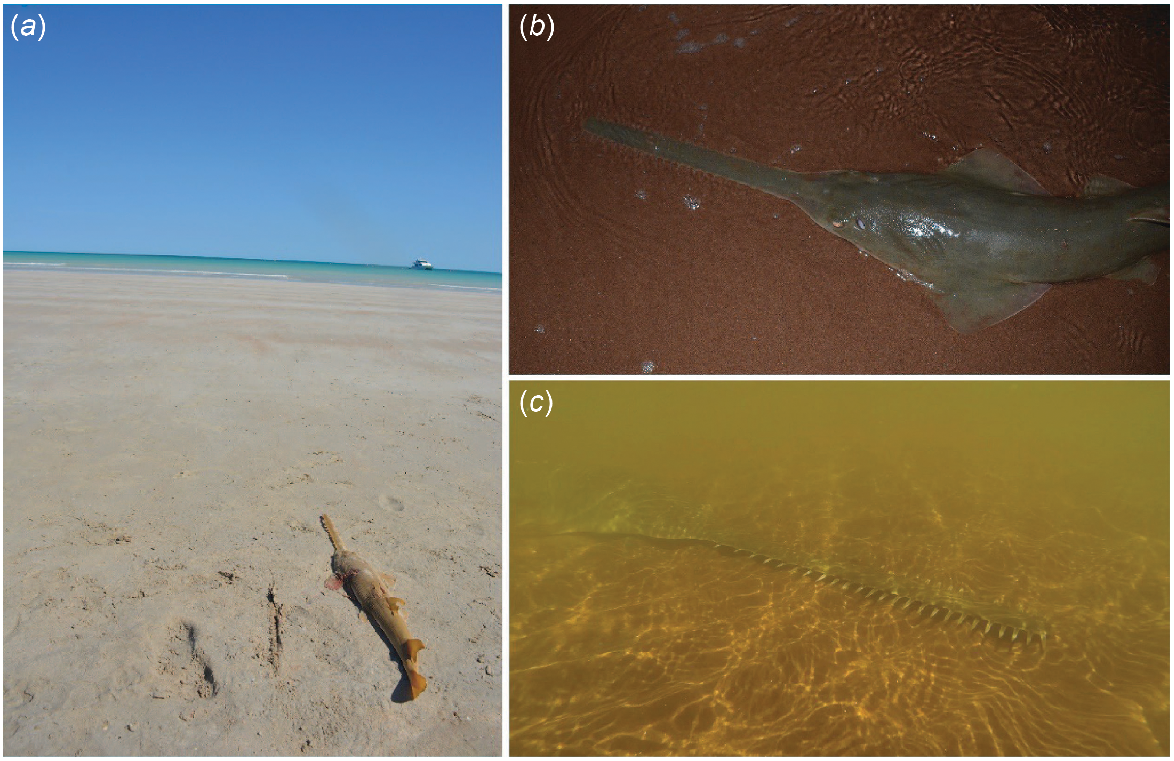
Images of individual green sawfish (Pristis zijsron) sampled for kinship investigations. (a) 2022 #38, young-of-the-year (YOY) P. zijsron (875-mm stretched total length, STL) sampled at Cable Beach, Broome, recently deceased, which was a half sibling of a sawfish captured in 2014 in the Ashburton River estuary. (b) 2014 #02, YOY P. zijsron (874 mm STL) sampled in the Ashburton River estuary, which was a half sibling of two full siblings captured at Cape Keraudren in 2008. (c) 2014 #05, ~2-year-old juvenile P. zijsron (1620 mm STL) sampled in the Ashburton River estuary, which was the half sibling of a sawfish captured at Cape Keraudren in 2008.
Dispersal
This is the first study to provide evidence of long-distance movement for the green sawfish, Pristis zijsron, which we define as any movement from one site to another where the distance between those sites exceeds 100 km. Telemetry data indicate that the movement of juvenile P. zijsron is generally confined to small core-activity spaces, concentrated in shallow waters, with home range increasing with growth (Morgan et al. 2017). Considering that all P. zijsron included in the kinship analyses were juveniles, the differences in capture locations between half siblings sampled >500 km apart are likely to be due to long-distance, parental movement between sites.
Previous genetic studies have reported that both female and male P. zijsron are regionally philopatric, with restricted gene flow at large spatial scales in northern Australia indicating limited dispersal in both sexes (Phillips et al. 2017; Ingelbrecht et al. 2024), although the degree to which philopatry occurs can vary among individuals or populations of the same species (Trenham et al. 2001). Furthermore, male-biased dispersal has been reported for the largetooth sawfish, Pristis pristis (Linnaeus, 1758), and the narrow sawfish, Anoxypristis cuspidata (Latham, 1794) (Phillips et al. 2017; Green et al. 2018), which may suggest that half siblings sampled more than 500 km apart are more likely to be related paternally. However, movement over similar distances is not exclusive to males for all Rhinopristiformes; Barnett et al. (2024) tracked a mature female giant shovelnose ray, Glaucostegus typus (Anonymous [Bennett], 1830) (~2490 mm TL), over 700 km south from its initial tagging location in Abbot Bay, Queensland. Thus, additional telemetry or genetic work is required to determine whether long-distance dispersal is sex-biased in P. zijsron, or related to reproduction or resource availability.
Pristis zijsron is legally protected within Australian Commonwealth waters under the Environment Protection and Biodiversity Conservation Act 1999, having been listed as Vulnerable in 2008 after satisfying Criterion 1 (i.e. decline in numbers) of the Environment Protection and Biodiversity Conservation Act eligibility requirements (Threatened Species Scientific Committee 2008; Department of the Environment 2015). In Western Australia, P. zijsron is also protected under the Wildlife Conservation Act 1950 and the Fish Resources Management Act 1994, which prohibits the take, possession, sale, purchase or consignment of P. zijsron and its products within the state (Department of the Environment 2015), although even with this protection, P. zijsron remains threatened by habitat degradation, gillnet and trawl fisheries in Australian waters (Kyne et al. 2021; Harry et al. 2024). A national recovery plan is, however, in place for sawfishes listed under the Environment Protection and Biodiversity Conservation Act, which includes a series of objectives and actions to improve outcomes for sawfishes (see Department of the Environment 2015). This work directly contributes to one of these objectives; specifically, objective 9b, which lists ‘the identification of important habitats for all life stages of sawfish species, including connectivity between regions’ as Priority 1 (i.e. prompt action necessary to mitigate key threats and provide valuable information to help identify long-term population trends) (Department of the Environment 2015). This study revealed the presence of half siblings between the Ashburton River and Cable Beach, thereby providing genetic evidence of movement and population connectivity between the Pilbara and Kimberley regions. Dispersal of individuals over large spatial scales reduces the risk of localised extinction for P. zijsron, as it suggests that assemblages could be replenished from elsewhere in the event of population decline. However, long-distance coastal movement of P. zijsron could be impeded, should major structures be established along the shoreline, with previous research demonstrating that movement of smaller juveniles between the Ashburton River and adjacent tidal creeks was constrained following construction of an offloading facility (see Lear et al. 2024). The potential impediment to long-distance movement should be taken into consideration in any coastal development plans. By providing important insight into the movement behaviour of P. zijsron, this research will contribute to more effective conservation strategies for this threatened species.
Data availability
The data that support this study are available from the corresponding author on reasonable request.
Declaration of funding
This study was supported by Murdoch University, Chevron Australia, the Western Australian Marine Science Institute, BCI Minerals, Leichhardt Salt, Protect Ningaloo, Cape Conservation Group and Minderoo Foundation.
Author contributions
Jack Ingelbrecht and David Morgan conceptualised the project and were responsible for the methods and investigation; Jack Ingelbrecht was responsible for formal analyses; all authors contributed to tissue sample collections, data curation, and writing, reviewing and editing the manuscript.
Acknowledgements
We thank the many people involved in this project, including Andrew Slater, Geoff Herbert, James Keleher, Mat Fraser, Paul de Lestang, Steve Moore, Jorma Nolan, Kathryn Dyball, Kevin Feldheim, Chloe Davidson, Stevie Sweeting, Colin Simpfendorfer and Patricia Charvet. We also thank the Chevron Indigenous Sea Rangers, Misty Shipway (DBCA), Colin Bartley and Aiden Mitchell for assistance with sample collections. We recognise and thank the Traditional Owners of the land on which this work took place.
References
Barnett A, Jaine FRA, Bierwagen SL, Lubitz N, Abrantes K, Heupel MR, Harcourt R, Huveneers C, Dwyer RG, Udyawer V, Simpfendorfer CA, Miller IB, Scott-Holland T, Kilpatrick CS, Williams SM, Smith D, Dudgeon CL, Hoey AS, Fitzpatrick R, Osborne FE, Smoothey AF, Butcher PA, Sheaves M, Fisher EE, Svaikauskas M, Ellis M, Kanno S, Cresswell BJ, Flint N, Armstrong AO, Townsend KA, Mitchell JD, Campbell M, Peddemors VM, Gustafson JA, Currey-Randall LM (2024) From little things big things grow: enhancement of an acoustic telemetry network to monitor broad-scale movements of marine species along Australia’s east coast. Movement Ecology 12, 31.
| Crossref | Google Scholar |
Carlson J, Blanco-Parra MP, Bonfil-Sanders R, Charles R, Charvet P, Chevis M, Dulvy NK, Espinoza M, Faria V, Ferretti F, Fordham S, Giovos I, Graham J, Grubbs D, Pacoureau N, Phillips NM (2022) Smalltooth sawfish Pristis pectinata. In ‘The IUCN Red List of Threatened Species 2022’. e.T18175A58298676. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/18175/58298676 [Verified 28 February 2024]
Compagno LJV, Last PR (1998) FAO species identification guide for fishery purposes: batoid fishes, chimaeras and bony fishes. In ‘The living marine resources of the Western Central Pacific’. (Eds KE Carpenter, VH Niem) pp. 1410–1417. (Food and Agriculture Organization of the United Nations: Rome, Italy)
Department of the Environment (2015) Sawfish and river sharks multispecies recovery plan. (Commonwealth of Australia) Available at https://www.dcceew.gov.au/sites/default/files/documents/sawfish-river-sharks-multispecies-recovery-plan.pdf [Verified 04 November 2024]
Dulvy NK, Davidson LNK, Kyne PM, Simpfendorfer CA, Harrison LR, Carlson JK, Fordham SV (2016) Ghosts of the coast: global extinction risk and conservation of sawfishes. Aquatic Conservation: Marine and Freshwater Ecosystems 26, 134-153.
| Crossref | Google Scholar |
Espinoza M, Bonfil-Sanders R, Carlson J, Charvet P, Chevis M, Dulvy NK, Everett B, Faria V, Ferretti F, Fordham S, Grant MI, Haque AB, Harry AV, Jabado RW, Jones GCA, Kelez S, Lear KO, Morgan DL, Phillips NM, Wueringer BE (2022) Largetooth sawfish Pristis pristis. In ‘The IUCN Red List of Threatened Species 2022’. e.T18584848A58336780. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/18584848/58336780 [Verified 28 February 2024]
Goudet J, Kay T, Weir BS (2018) How to estimate kinship. Molecular Ecology 27, 4121-4135.
| Crossref | Google Scholar | PubMed |
Grant MI, Charles R, Fordham S, Harry AV, Lear KO, Morgan DL, Phillips NM, Simeon B, Wakhida Y, Wueringer BE (2022) Dwarf sawfish Pristis clavata. In ‘The IUCN Red List of Threatened Species 2022’. e.T39390A68641215. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/39390/68641215 [Verified 28 February 2024]
Green ME, D’Anastasi BR, Hobbs JPA, Feldheim K, McAuley R, Peverell S, Stapley J, Johnson G, Appleyard SA, White WT, Simpfendorfer CA, van Herwerden L (2018) Mixed-marker approach suggests maternal philopatry and sex-biased behaviours of narrow sawfish Anoxypristis cuspidata. Endangered Species Research 37, 45-54.
| Crossref | Google Scholar |
Gruber B, Unmack PJ, Berry OF, Georges A (2018) DARTR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources 18, 691-699.
| Crossref | Google Scholar | PubMed |
Haque AB, Charles R, D’Anastasi B, Dulvy NK, Faria V, Fordham S, Grant MI, Harry AV, Jabado RW, Lear KO, Morgan DL, Tanna A, Wakhida Y, Wueringer BE (2023) Narrow sawfish Anoxypristis cuspidata. In ‘The IUCN Red List of Threatened Species 2023’. e.T39389A58304073. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/39389/58304073 [Verified 28 February 2024]
Harry AV, Everett B, Faria V, Fordham S, Grant MI, Haque AB, Ho H, Jabado RW, Jones GCA, Lear KO, Morgan DL, Phillips NM, Spaet JLY, Tanna A, Wueringer BE (2022) Green sawfish Pristis zijsron. In ‘The IUCN Red List of Threatened Species’. e.T39393A58304631. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/39393/58304631 [Verified 28 February 2024]
Harry AV, Wakefield CB, Newman SJ, Braccini JM (2024) Trends in catch rates of sawfish on the Australian North West Shelf. Endangered Species Research 53, 23-33.
| Crossref | Google Scholar |
Huisman J (2017) Pedigree reconstruction from SNP data: parentage assignment, sibship clustering and beyond. Molecular Ecology Resources 17, 1009-1024.
| Crossref | Google Scholar | PubMed |
Ingelbrecht J, Lear KO, Phillips NM, Wueringer BE, Lymbery AJ, Norman BM, Morgan DL (2024) Kinship assessment and insights into reproductive behaviour of the Critically Endangered green sawfish Pristis zijsron in Western Australia. Aquatic Conservation: Marine and Freshwater Ecosystems 34, e4213.
| Crossref | Google Scholar |
Jones OR, Wang J (2010a) Molecular marker-based pedigrees for animal conservation biologists. Animal Conservation 13, 26-34.
| Crossref | Google Scholar |
Jones OR, Wang J (2010b) COLONY: a program for parentage and sibship inference from multilocus genotype data. Molecular Ecology Resources 10, 551-555.
| Crossref | Google Scholar | PubMed |
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. In ‘Data production and analysis in population genomics. Vol. 888’. Methods in Molecular Biology. (Eds F Pompanon, A Bonin) pp. 67–89. (Humana Press) doi:10.1007/978-1-61779-870-2_5
Lear KO, Fazeldean T, Bateman RL, Inglebrecht J, Morgan DL (2023) Growth and morphology of Critically Endangered green sawfish Pristis zijsron in globally important nursery habitats. Marine Biology 170, 70.
| Crossref | Google Scholar |
Lear KO, Ebner BC, Fazeldean T, Bateman RL, Morgan DL (2024) Effects of coastal development on sawfish movements and the need for marine animal crossing solutions. Conservation Biology 38, e14263.
| Crossref | Google Scholar |
Mace GM, Collar NJ, Gaston KJ, Hilton-Taylor C, Akcakaya HR, Leader-Williams N, Milner-Gulland EJ, Stuart SN (2008) Quantification of extinction risk: IUCN’s system for classifying threatened species. Conservation Biology 22, 1424-1442.
| Crossref | Google Scholar | PubMed |
Madsen T, Stille B, Shine R (1996) Inbreeding depression in an isolated population of adders Vipera berus. Biological Conservation 75, 113-118.
| Crossref | Google Scholar |
Morgan DL, Whitty JM, Phillips NM, Thorburn DC, Chaplin JA, McAuley R (2011) North-western Australia as a hotspot for endangered elasmobranchs with particular reference to sawfishes and the Northern River Shark. Journal of the Royal Society of Western Australia 94, 345-358.
| Google Scholar |
Morgan DL, Allen MG, Ebner BC, Whitty JM, Beatty SJ (2015) Discovery of a pupping site and nursery for critically endangered green sawfish Pristis zijsron. Journal of Fish Biology 86, 1658-1663.
| Crossref | Google Scholar | PubMed |
Morgan DL, Ebner BC, Allen MG, Gleiss AC, Beatty SJ, Whitty JM (2017) Habitat use and site fidelity of neonate and juvenile green sawfish Pristis zijsron in a nursery area in Western Australia. Endangered Species Research 34, 235-249.
| Crossref | Google Scholar |
Peverell SC (2005) Distribution of sawfishes (Pristidae) in the Queensland Gulf of Carpentaria, Australia, with notes on sawfish ecology. Environmental Biology of Fishes 73, 391-402.
| Crossref | Google Scholar |
Phillips NM, Chaplin JA, Peverell SC, Morgan DL (2017) Contrasting population structures of three Pristis sawfishes with different patterns of habitat use. Marine and Freshwater Research 68, 452-460.
| Crossref | Google Scholar |
Premachandra HKA, Nguyen NH, Knibb W (2019) Effectiveness of SNPs for parentage and sibship assessment in polygamous yellowtail kingfish Seriola lalandi. Aquaculture 499, 24-31.
| Crossref | Google Scholar |
Speed D, Balding DJ (2015) Relatedness in the post-genomic era: is it still useful? Nature Reviews Genetics 16, 33-44.
| Crossref | Google Scholar | PubMed |
Threatened Species Scientific Committee (2008) Listing advice for Pristis zijsron (green sawfish). (TSSC) Available at http://www.environment.gov.au/biodiversity/threatened/species/pubs/68442-listing-advice.pdf [Verified 04 November 2024]
Trenham PC, Koenig WD, Shaffer HB (2001) Spatially autocorrelated demography and interpond dispersal in the salamander Ambystoma californiense. Ecology 82, 3519-3530.
| Crossref | Google Scholar |


