Insights into the global emergence of antifungal drug resistance
Kylie Boyce A D , Orla Morrissey B , Alexander Idnurm C and Ian Macreadie AA School of Science, RMIT University, Bundoora, Vic. 3083, Australia
B Department of Infectious Diseases, The Alfred Hospital, Melbourne, Vic. 3004, Australia
C School of BioSciences, The University of Melbourne, Parkville, Vic. 3010, Australia
D Tel: +61 3 9925 7101, Email: kylie.boyce@rmit.edu.au
Microbiology Australia 40(2) 87-91 https://doi.org/10.1071/MA19024
Published: 18 April 2019
The global prevalence of fungal diseases has escalated in the last several decades. Currently, it is estimated that fungi infect 1.7 billion people annually and result in 1.5 million deaths every year1. Deaths due to fungal infections are increasing, with mortality often exceeding 50%, further increasing to 100% if treatment is delayed1. Despite these staggering figures, the contribution of fungal infections to the global burden of disease remains under-recognised. In Australia, over a 5-year period fungal infections cost Australia an estimated $583 million2. The median cost for one invasive fungal disease (IFD) is AU$30 957, increasing to AU$80 291 if the patient is admitted to an intensive care unit3. Treatment of fungal infections poses significant challenges due to the small number of safe and effective antifungal drugs available and emerging antifungal drug resistance. Resistance to every class of antifungal drugs has been described and for some drug classes is extremely common4,5.
More than 90% of all reported fungal deaths result from species that belong to one of three genera: Candida, Aspergillus and Cryptococcus1. Candida species normally live on the epithelial surfaces of the host and are common in immunocompromised patients who have undergone invasive procedures and can form biofilms on medical devices1. Candida albicans is the most prevalent species causing disease, followed by C. glabrata, C. tropicalis, C. parapsilosis and C. krusei (recently renamed C. acidothermophilum)4. Candida auris is an emerging fungal pathogen that has intrinsic multi-drug resistance. Since first emerging in 2009, C. auris has spread across five continents, including Australia, and resulted in a number of nosocomial outbreaks6,7. Cryptococcosis, caused by Cryptococcus neoformans and Cryptococcus gattii, is the most common invasive fungal disease globally, at one point with an estimated >1M life-threatening infections per year1. C. neoformans, isolated from soil or bird excrement, has a worldwide distribution and predominantly causes infections in immunocompromised individuals. C. gattii is isolated from eucalyptus and other trees, so has a more restricted distribution, and causes infections largely in immunocompetent individuals8. Aspergillus species commonly cause invasive pulmonary aspergillosis in neutropenic patients, transplant recipients and in patients on immunosuppression (e.g. corticosteroids). Aspergillus species are found worldwide and are common in the environment resulting in continuous lung exposure1. The most common species are in the A. fumigatus complex, but A. flavus, A. niger, A. terreus and A. nidulans also cause infections in humans.
Antifungal drugs and emerging resistance
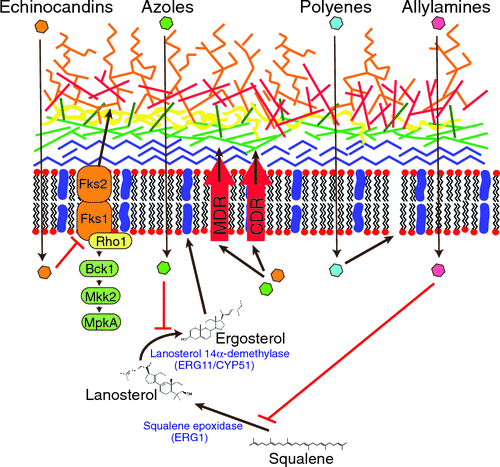
There are four main classes of antifungal drugs: polyenes, azoles, allylamines and echinocandins (Figure 1, Table 1). Polyenes (e.g. amphotericin B) bind ergosterol in the cell membrane and induce pore formation. Azoles (e.g. fluconazole, voriconazole) and allylamines (e.g. terbinafine) inhibit the enzymes lanosterol 14 α-demethylase (ERG11p/cyp51) and squalene epoxidase, respectively, which are needed for ergosterol biosynthesis in the fungal cell membrane. Echinocandins (e.g. caspofungin) inhibit the synthesis of glucan in the fungal cell wall. Echinocandins have few side-effects but are poorly absorbed and have limited efficacy (e.g. none against C. neoformans)22. Closely related species can exhibit differences in intrinsic susceptibility. For example, 98% of C. albicans isolates are susceptible to fluconazole whereas only 9% of C. krusei are susceptible4. Resistance to every family of antifungal drugs has been described and is at least partially responsible for the poor outcomes and high morbidity seen in patients with IFD. Resistance to azoles is common and increasing5.
|

|
The prevalence of antifungal resistance in Candida is not clear, mainly because of the differences in the levels of intrinsic susceptibilities between Candida species. Primary resistance to fluconazole is rare for C. albicans (1.4%), C. parapsilosis (3.6%) and C. tropicalis (4.1%) but species such as C. krusei are intrinsically resistant to fluconazole (78.3%)13. In a recent United States retrospective study acquired fluconazole resistance was reported in 19% of C. albicans infections13. At present, resistance to azoles is uncommon in Australia but may be increasing; a recent review of candidemia in Australia showed 16.7% of C. tropicalis isolates are resistant to both fluconazole and voriconazole23. A major concern is the emergence of infections caused by Candida species other than C. albicans, such as C. auris that has intrinsic multi-drug resistance. Currently C. auris is not established in Australia: the few Australian cases have come about through people being infected overseas (Victoria State Government, August 2018)7. Vigilance is warranted towards preventing this new multi-drug resistant pathogen from establishing a foothold in Australia.
A recent systematic review of resistance in Cryptococcus species from 1988–2017 revealed 10.6% of clinical isolates are fluconazole resistant and this rises to 24.1% in patients with relapsed cryptococcal infection24.
The first few instances of azole resistance in A. fumigatus were reported in the USA and Europe in the late 1980s and 90s in association with long-term antifungal treatment (extensively reviewed by Meis et al.25). Since then, the prevalence of resistant isolates has increased and has been found in patients who have not received azole treatment. The acquisition of resistance in patients who have not received azole treatment has led to the hypothesis that resistance can also be gained from the agricultural use of azoles17. In 1999, the prevalence of azole-resistant A. fumigatus in The Netherlands was 12.8% but current incidence is 20% and resistance has now been reported in 11 different countries5,26.
Molecular mechanisms of antifungal drug resistance
The emergence of antifungal drug resistance can occur due to changes in the genome9–11. On exposure to azole antifungal drugs, fungal pathogens can undergo a process termed heteroresistance in which rapid, yet reversible, resistance is conferred by the development of one or more aneuploidies (large scale chromosomal rearrangements) (reviewed by Morrow and Fraser12). Cells return to normal ploidy when the azole is removed, as the aneuploidies result in strains with reduced fitness both in culture and in the host10. In C. albicans, the most commonly occurring aneuploidy is an isochromosome of the left arm of chromosome 5, i(5L). This region contains the ERG11 gene encoding the target of fluconazole, as well as the TAC1 gene encoding the transcriptional activator of drug efflux genes9. In C. neoformans, chromosome 1, containing ERG11 and the azole transporter AFR1, is duplicated in response to increasing concentrations of fluconazole, followed by the successive duplication of chromosomes 4, 10 and 1410.
Resistance to azole drugs in C. albicans, C. parapsilosis, C. krusei, C. tropicalis,C. neoformans and A. fumigatus can also be acquired by single nucleotide mutations in the ERG11 gene (cyp51) encoding the target enzyme 14 α-demethylase14,15. Mutations that lead to overexpression of the efflux pumps encoded by MDR (major facilitator superfamily pumps) in C.albicans and C. parapsilosis or CDR genes (ATP-binding cassette pumps) also confer resistance in C. albicans, C. glabrata and C. krusei4. Mutations in C. glabrataPDR1, a transcription factor regulating the expression of drug efflux pumps, also confers resistance to fluconazole19. C. auris clinical isolates possess acquired mutations in ERG11 (azole resistance), FKS1 (echinocandin resistance) and FUR1 (5-flucytosine resistance)6,27. Candida species become resistant to echinocandins due to mutations in the FKS genes encoding the two subunits of the target enzyme 1,3-ß-D-glucan synthase, specifically FKS1 in C. albicans, C. krusei and C. tropicalis and both FKS1 and FKS2 in C. glabrata5.
In A. fumigatus, the most common mechanism of azole resistance occurring within a host involves mutations in the cyp51A (ERG11) gene, which prevent the azole from binding to the heme molecule16,25. In contrast, A. fumigatus resistant clinical strains acquired from the environment most commonly possess a duplication of a 34 bp tandem repeat in the promoter region of cyp51A, combined with a specific substitution that causes overexpression of the gene5,18. These mutations confer multi-drug resistance and have been found to be correlated with exposure to agricultural azoles in the environment16,17.
In comparison to resistance to azoles, almost nothing is known about the mechanisms that give rise to resistance to polyenes in fungi. Strains naturally resistant to amphotericin B exist but resistance can also develop on treatment. In Candida species, acquisition of resistance to amphotericin B can occur through mutations of some of the ERG genes of the ergosterol biosynthesis pathway, although most resistant strains have only been characterised by biochemical analysis of membrane sterol composition. The only C. neoformans amphotericin B resistant isolate characterised had a mutation in ERG2, encoding sterol 8-7 isomerase21.
The role of mutation rate in accelerating the emergence of resistance
Recent studies in C. glabrata and C. neoformans have revealed that a proportion of clinical isolates possess an elevated mutation rate (a ‘mutator’ phenotype), which can contribute to the rapid emergence of spontaneous antifungal drug resistance via increasing the opportunity for selectively advantageous mutations to occur28,29. Initial studies showed that 55% of C. glabrata clinical isolates contain non-synonymous variation in the MSH2 gene, which encodes a component of mismatch DNA repair28. The presence of non-synonymous variation in MSH2 correlated with multi-drug resistance28. C. glabrata clinical isolates possessing non-synonymous variation in MSH2 have now been detected in clinical populations in many parts of the world with varying prevalence (North America 55%; India 69%; France 44%; Korea 65%)30–32. However, there is not always an obvious correlation with drug resistance. In addition, an assessment of mutation rate was not performed in these studies, leading to the criticism that the variation in MSH2 may not result in a true mutator phenotype. A recent new green fluorescent protein (GFP) reporter coupled with Fluorescence-Activated Cell Sorting (FACS) technique has been developed to test mutation rates in C. glabrata clinical isolates strains with different MSH2 alleles33. An elevated rate was not observed for isolates with the MSH2E231G/L269F allelic variant suggesting that not all non-synonymous variation in MSH2 results in a true mutator phenotype33. Clinical isolates of C. neoformans with non-synonymous variation in MSH2 have also been identified29,34. These isolates exhibit a mutator phenotype and an increase in the emergence of spontaneous fluconazole and amphotericin B resistance29. Deletion of MSH2 in both C. glabrata and C. neoformans results in high levels of spontaneous resistance to multiple types of antifungals28,29.
Conclusions
Resistance to antifungal drugs is clearly becoming an important clinical issue that will escalate in the future unless new classes of antifungals are developed. Early treatment strategies such as prophylaxis or extensive, long-term use of antifungals to avoid relapse frequently selects for drug resistance, as does environmental exposure possibly to agricultural azoles and over the counter use (e.g. fluconazole pessaries for vaginal thrush). An improved understanding of factors influencing the emergence of, and mechanisms of, resistance is required to develop effective future treatment strategies.
Conflicts of interest
Dr Morrissey has been a member of advisory boards for, received investigator-initiated grants from, and given lectures for Gilead Sciences, and Merck, Sharp and Dohme. All funds received are administered by Alfred Health/Monash University.
Acknowledgements
This research did not receive any specific funding.
References
[1] Brown, G.D. et al. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv13.| Hidden killers: human fungal infections.Crossref | GoogleScholarGoogle Scholar | 23253612PubMed |
[2] Slavin, M. et al. (2004) Burden of hospitalization of patients with Candida and Aspergillus infections in Australia. Int. J. Infect. Dis. 8, 111–120.
| Burden of hospitalization of patients with Candida and Aspergillus infections in Australia.Crossref | GoogleScholarGoogle Scholar | 14732329PubMed |
[3] Ananda-Rajah, M.R. et al. (2011) Attributable hospital cost and antifungal treatment of invasive fungal diseases in high-risk hematology patients: an economic modeling approach. Antimicrob. Agents Chemother. 55, 1953–1960.
| Attributable hospital cost and antifungal treatment of invasive fungal diseases in high-risk hematology patients: an economic modeling approach.Crossref | GoogleScholarGoogle Scholar | 21357302PubMed |
[4] Beardsley, J. et al. (2018) Responding to the emergence of antifungal drug resistance: perspectives from the bench and the bedside. Future Microbiol. 13, 1175–1191.
| Responding to the emergence of antifungal drug resistance: perspectives from the bench and the bedside.Crossref | GoogleScholarGoogle Scholar | 30113223PubMed |
[5] Gamaletsou, M.N. et al. (2018) Invasive fungal infections in patients with hematological malignancies: Emergence of resistant pathogens and new antifungal therapies. Turk. J. Haematol. 35, 1–11.
| Invasive fungal infections in patients with hematological malignancies: Emergence of resistant pathogens and new antifungal therapies.Crossref | GoogleScholarGoogle Scholar | 29391334PubMed |
[6] Rhodes, J. et al. (2018) Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 7, 43.
| 29593275PubMed |
[7] Heath, C.H. et al. (2019) Candida auris sternal osteomyelitis in a man from Kenya visiting Australia, 2015. Emerg. Infect. Dis. 25, 192–194.
| Candida auris sternal osteomyelitis in a man from Kenya visiting Australia, 2015.Crossref | GoogleScholarGoogle Scholar | 30561310PubMed |
[8] Slavin, M.A. and Chakrabarti, A. (2012) Opportunistic fungal infections in the Asia-Pacific region. Med. Mycol. 50, 18–25.
| Opportunistic fungal infections in the Asia-Pacific region.Crossref | GoogleScholarGoogle Scholar | 21905945PubMed |
[9] Selmecki, A. et al. (2008) An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68, 624–641.
| An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1.Crossref | GoogleScholarGoogle Scholar | 18363649PubMed |
[10] Sionov, E. et al. (2010) Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6, e1000848.
| Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes.Crossref | GoogleScholarGoogle Scholar | 20368972PubMed |
[11] Almeida, A.M. et al. (2007) Molecular typing and antifungal susceptibility of clinical sequential isolates of Cryptococcus neoformans from Sao Paulo State, Brazil. FEMS Yeast Res. 7, 152–164.
| Molecular typing and antifungal susceptibility of clinical sequential isolates of Cryptococcus neoformans from Sao Paulo State, Brazil.Crossref | GoogleScholarGoogle Scholar | 17311593PubMed |
[12] Morrow, C.A. and Fraser, J.A. (2013) Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin. Cell Dev. Biol. 24, 339–346.
| Ploidy variation as an adaptive mechanism in human pathogenic fungi.Crossref | GoogleScholarGoogle Scholar | 23380396PubMed |
[13] Sanguinetti, M. et al. (2015) Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 58, 2–13.
| Antifungal drug resistance among Candida species: mechanisms and clinical impact.Crossref | GoogleScholarGoogle Scholar | 26033251PubMed |
[14] Rodero, L. et al. (2003) G484S amino acid substitution in lanosterol 14-α demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47, 3653–3656.
| G484S amino acid substitution in lanosterol 14-α demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate.Crossref | GoogleScholarGoogle Scholar | 14576140PubMed |
[15] Morio, F. et al. (2010) Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66, 373–384.
| Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature.Crossref | GoogleScholarGoogle Scholar | 20226328PubMed |
[16] Chowdhary, A. et al. (2014) Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. J. Antimicrob. Chemother. 69, 555–557.
| Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India.Crossref | GoogleScholarGoogle Scholar | 24084639PubMed |
[17] Chowdhary, A. et al. (2013) Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 9, e1003633.
| Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health.Crossref | GoogleScholarGoogle Scholar | 24204249PubMed |
[18] Gsaller, F. et al. (2016) Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog. 12, e1005775.
| Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex.Crossref | GoogleScholarGoogle Scholar | 27973537PubMed |
[19] Whaley, S.G. and Rogers, P.D. (2016) Azole resistance in Candida glabrata. Curr. Infect. Dis. Rep. 18, 41.
| Azole resistance in Candida glabrata.Crossref | GoogleScholarGoogle Scholar | 27761779PubMed |
[20] Yamada, T. et al. (2017) Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob. Agents Chemother. 61, e00115-17.
| Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene.Crossref | GoogleScholarGoogle Scholar | 28416557PubMed |
[21] Kelly, S.L. et al. (1994) Resistance to amphotericin B associated with defective sterol delta 8-->7 isomerase in a Cryptococcus neoformans strain from an AIDS patient. FEMS Microbiol. Lett. 122, 39–42.
| Resistance to amphotericin B associated with defective sterol delta 8-->7 isomerase in a Cryptococcus neoformans strain from an AIDS patient.Crossref | GoogleScholarGoogle Scholar | 7958776PubMed |
[22] Maligie, M.A. and Selitrennikoff, C.P. (2005) Cryptococcus neoformans resistance to echinocandins: (1,3)β-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 49, 2851–2856.
| Cryptococcus neoformans resistance to echinocandins: (1,3)β-glucan synthase activity is sensitive to echinocandins.Crossref | GoogleScholarGoogle Scholar | 15980360PubMed |
[23] Chapman, B. et al. (2017) Changing epidemiology of candidaemia in Australia. J. Antimicrob. Chemother. 72, 1270.
| Changing epidemiology of candidaemia in Australia.Crossref | GoogleScholarGoogle Scholar | 28204502PubMed |
[24] Bongomin, F. et al. (2017) Global and multi-national prevalence of fungal diseases—estimate precision. J. Fungi (Basel) 3, 57.
| Global and multi-national prevalence of fungal diseases—estimate precision.Crossref | GoogleScholarGoogle Scholar |
[25] Meis, J.F. et al. (2016) Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150460..
| 28080986PubMed |
[26] Verweij, P.E. et al. (2009) Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect. Dis. 9, 789–795.
| Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use?Crossref | GoogleScholarGoogle Scholar | 19926038PubMed |
[27] Bidaud, A.L. et al. (2018) Candida auris: an emerging drug resistant yeast – a mini-review. J. Mycol. Med. 28, 568–573.
| Candida auris: an emerging drug resistant yeast – a mini-review.Crossref | GoogleScholarGoogle Scholar | 30030072PubMed |
[28] Healey, K.R. et al. (2016) Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 7, 11128.
| Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance.Crossref | GoogleScholarGoogle Scholar | 27020939PubMed |
[29] Boyce, K.J. et al. (2017) Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. MBio 8, e00595-17.
| Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans.Crossref | GoogleScholarGoogle Scholar | 28559486PubMed |
[30] Singh, A. et al. (2018) Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob. Agents Chemother. 62, e00195-e18.
| Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway.Crossref | GoogleScholarGoogle Scholar | 30082281PubMed |
[31] Dellière, S. et al. (2016) Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a French cohort of patients harboring low rates of resistance. Front. Microbiol. 7, 2038.
| Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a French cohort of patients harboring low rates of resistance.Crossref | GoogleScholarGoogle Scholar | 28066361PubMed |
[32] Byun, S.A. et al. (2018) Multilocus sequence typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: association with antifungal resistance, mutations in mismatch repair gene (Msh2), and clinical outcomes. Front. Microbiol. 9, 1523.
| Multilocus sequence typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: association with antifungal resistance, mutations in mismatch repair gene (Msh2), and clinical outcomes.Crossref | GoogleScholarGoogle Scholar | 30057573PubMed |
[33] Shor, E. et al. (2019) A novel, drug resistance-independent, fluorescence-based approach to measure mutation rates in microbial pathogens. MBio 10, e00120-19.
| A novel, drug resistance-independent, fluorescence-based approach to measure mutation rates in microbial pathogens.Crossref | GoogleScholarGoogle Scholar | 30808701PubMed |
[34] Rhodes, J. et al. (2017) A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection. G3 (Bethesda) 7, 1165–1176.
| A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection.Crossref | GoogleScholarGoogle Scholar | 28188180PubMed |
Biographies
Dr Kylie Boyce is an expert in the molecular genetics of pathogenic fungi. Her research at RMIT University focuses on how pathogenic fungi interact with, and adapt to, the human host. She has recently been investigating how DNA repair and mutation rate contribute to the microevolution of fungal pathogens and their ability to rapidly generate spontaneous antifungal drug resistance.
Dr Orla Morrissey is an Infectious Diseases Physician at Alfred Health, Melbourne and Senior Lecturer in the Department of Infectious Diseases at Monash University, Melbourne. Dr Morrissey is a lead clinician within the Immunocompromised Host Consult Service at Alfred Health and co-chair of the Australia and New Zealand Mycoses Interest Group. Dr Morrissey is active in the research sphere: determining the epidemiology of a variety of opportunistic infections; determining Aspergillus virulence factors and examining inflammatory responses to Aspergillus.
Dr Alexander Idnurm is an expert in human and agricultural pathogenic fungi at Melbourne University. His research is focused on how fungi respond to their environment to change physiology and development. His research encompasses the genetic and molecular biology analyses of a number of different fungal species, providing an ability to take comparative approaches across the fungal kingdom. A recent focus has been on how quickly fungi change during their encounters with hosts, as this microevolution has ramifications for the emergence of antifungal drug resistance.
Prof Ian Macreadie is a molecular microbiologist who works with yeast as a model for studying Alzheimer’s disease. He also studies the effects of biochemicals and drugs on yeast, as well as studying the drug resistance of yeast. He teaches Industrial Microbiology at RMIT University and leads students to learn about how the gut microbiota of Australian animals aids their survival.


