The diminished antimicrobial pipeline
Mark AT BlaskovichInstitute for Molecular Bioscience, The University of Queensland, Brisbane, Qld 4072, Australia
Email: m.blaskovich@uq.edu.au
Microbiology Australia 40(2) 92-96 https://doi.org/10.1071/MA19025
Published: 18 April 2019
Australians love antibiotics, with one of the highest rates of human antibiotic usage in the world. Unfortunately, they are being loved to death, as high rates of inappropriate use, both here and around the globe, are contributing to the rise of drug-resistant bacteria against which our current arsenal of antibiotics is becoming increasingly ineffective. In the past, advancements in developing new antibiotics kept pace with developing resistance, but we are now facing a deadly reality where the pipeline of ‘new and improved’ antibiotics is rapidly drying up. There are a number of global initiatives attempting to reprime the pipeline, but the exit of major pharmaceutical companies from antibiotic research and the poor financial performance of antibiotic-focused biotechnology companies continues.
Antimicrobial resistance (AMR) is becoming an increasing threat to human health, with the World Health Organization (WHO), the United Nations, and the G7 Health Ministers all releasing statements recognising the threat posed by antimicrobial resistance, and the potential to enter a post-antibiotic era.
Resistance against antibiotics arises in a number of ways. The antibiotic can be prevented from reaching its site of action inside the microorganism, either by blocking entry (modifications to the cell wall or porins), or by increasing active export via upregulation of efflux pumps. The antibiotic’s target can be modified to make the interaction less effective (e.g. alteration of Lipid A structure causing polymyxin resistance, or mutations to the ribosome binding site generating resistance to macrolides). The antibiotic itself can be modified or deactivated (e.g. β-lactamase cleavage of lactam antibiotics, or acetylation of aminoglycoside antibiotics). Antibiotic resistance is nothing new, with clinical resistance detected against EVERY antibiotic ever introduced, generally within a few years of its first human use. Indeed, genes encoding resistance to β-lactam, tetracycline and glycopeptide antibiotics have been isolated from 30 000 year old permafrost1.
Antibiotics are incredibly valuable as they one of the few medicines that actually cures the disease it treats. Unfortunately, their widespread misuse is the predominant driver of resistance. Approximately two-thirds of the world’s antibiotics are used in animals2, and of the remaining one-third, about two-thirds are inappropriately prescribed2, often for viral-caused colds and flus where antibiotics have no effect. Unfortunately, the general perception that antibiotics are safe, with no drawbacks to prescribing/taking them if they are not needed, helps to drive this overuse. Like most drugs, antibiotics do have toxic side-effects, particularly kidney and muscle damage. Increasingly, less obvious detrimental effects are being identified, such as changes in the microbiome that are associated with obesity, cancer, inflammatory bowel disease, arthritis and autism3. Unfortunately, both human4 and animal5 use of antibiotics is still increasing.
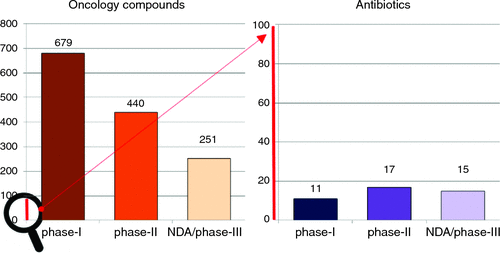
So why do we now have concerns over the scarcity of new antibiotics? Since the 1980s almost all major pharmaceutical companies have exited from antibiotic discovery, with one of the last, Novartis, announcing their exit in July 2018. Even ‘successful’ biotech companies are struggling: Achaogen’s first drug, plazomicin, was approved in June 2018, but within weeks it had laid off 28% of its staff, with further restructuring in November, and a plummeting stock price(from over $12 before approval to less than $1 in March 2019). The lack of antibiotic development is primarily driven by financial reasons, with similar development costs as other classes of therapeutics, but substantially lower potential returns. For example, sales for the first two years for two of the more successful recent antibiotics, Teflaro (US$50m) and Avycaz (US$80m) are dwarfed by those of the anti-diabetes drug Januvia (US$1400m) and the anti-epileptic/anti-anxiety drug Lyrica (US$1300m)6. Most infections can still be treated by generic antibiotics that usually cost less than US$100 for a course of treatment, and this apparently limits the palatability for what can be charged for the best new antibiotics to US$1000/day (or less than $15 000 for a standard two-week treatment). In contrast, an anti-cancer therapy, such as the newest CAR-T technology, can charge over US$400 000 for a single treatment. Other medications must be taken for a lifetime. These situations are much more appealing to investors and shareholders. This has led to our current crisis, with multiple reports7–9 documenting that the clinical antibiotic pipeline contains less than 50 candidates. In stark contrast, there were over 800 potential new anti-cancer drugs in 201510. Alarmingly, instead of the traditional funnel of attrition through the three stages of clinical testing (approximately 50% fewer candidates in Phase 2 than Phase 1, and another 50% fewer in Phase 3), the antibiotic pipeline is flat, if not reversed (Figure 1). Given the length of time it takes to progress from Phase 1 to an approved drug (often 10 years), even if we immediately replenished the start of the pipeline, there’s an enormous gap before they become available to physicians.
|
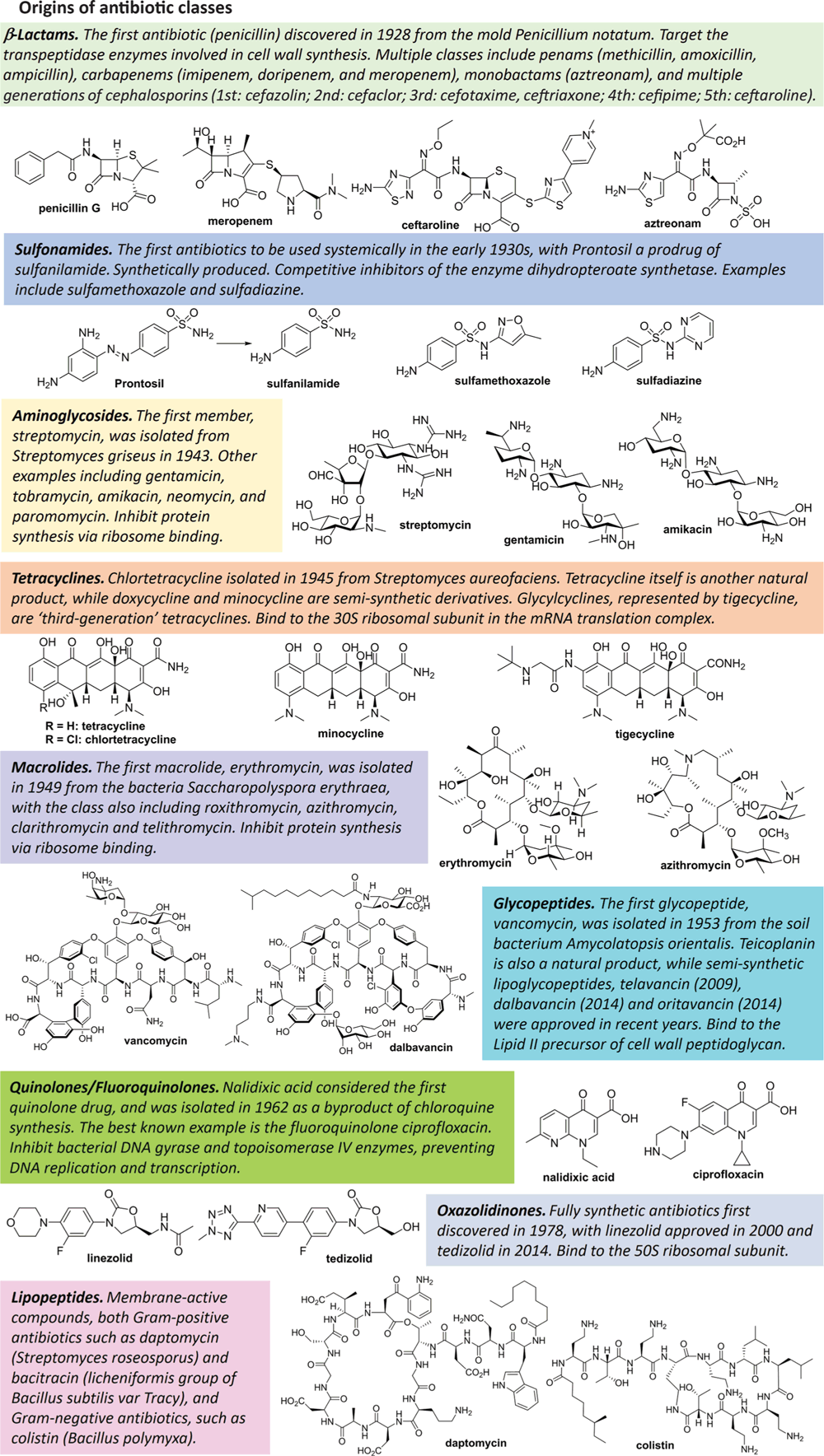
If we are able to overcome the financial hurdles, how do we actually go about discovering new antibiotics? Ironically, microbiology has played a pivotal role in fighting back against microbial infections. Almost all antibiotic classes are originally derived from bacteria or fungi, particular actinomycetes11 (Figure 2), and some (e.g. vancomycin, polymyxin) are still produced industrially by fermentation. Subsequent generations of many antibiotics are ‘semisynthetic’, where chemical modifications are conducted on microbially produced complex starting materials. Others, such as lactam antibiotics and fluoroquinolones, are now completely chemically synthesised.
After the ‘golden age’ of antibiotic discovery (1940s–1970s), natural product isolation of new antibiotics showed diminishing returns, with screening programs generally rediscovering known antibiotics or minor variations. Instead, most new antibiotics resulted from optimisation of existing classes, employing rational design to overcome resistance mechanisms and improve potency or pharmacokinetic properties. The 1980s–1990s also focused on combining high-throughput screening of combinatorial libraries with target-based discovery, relying on advances in genomics that identified new bacterial targets distinct from human analogues. Unfortunately, this approach was largely unsuccessful, as highlighted by summaries of research programs at GlaxoSmithKline12 and Astrazeneca13. Good potency at the desired isolated in vitro target was usually lost when moving to phenotypic testing due to the many mechanisms bacteria have evolved to keep compounds out. However, recent advances in the development of inhibitors of LpxC14, a key enzyme in the synthesis of the Gram-negative membrane constituents, demonstrate that target-based research can be a viable approach to produce clinical candidates. Future programs will likely have improved success, guided by predictive rules to design compounds able to penetrate into Gram-negative bacteria15.
Recent years have seen a renaissance in natural product antibiotic discovery, applying new techniques that overcome the limitations of traditional approaches. Standard culture conditions only grow a fraction of the microorganisms present in an environmental sample, so new methods culture ‘hidden’ bacteria and fungi to identify unique compounds. The novel antibiotic teixobactin was isolated via an ‘Ichip’ that allows for the environmental bacterium to grow in situ16. Researchers are also testing bacteria that grow in unusual places and produce unusual metabolites, such as deep ocean marine bacteria, thermal vent extremophiles, high-salt halophiles or bacteria found in caves17. Co-culturing microorganisms together can force their defense mechanisms to produce new metabolites that normally would not be identified18, as can addition of chemical additives that similarly stimulate the defense mechanism19. Finally, metagenomic sequencing approaches can identify biosynthetic gene clusters from individual microorganisms or environmental communities, with the sequences used to predict the structures of new compounds20, as demonstrated by the culture-independent discovery of the malacidins21.
However, new antibiotics are not the only way to fight antimicrobial resistance22. ‘Resistance breakers’, adjuvants that work in combination with an antibiotic to overcome resistance, have long been used with β-lactam antibiotics to inhibit bacterial β-lactamase enzymes from destroying the lactam. Companies such as Spero Therapeutics are now developing other compounds that can expand the spectrum and enhance the potency of other existing antibiotics. Vaccines have been incredibly effective at reducing meningococcal and pneumococcal infections, and much research is ongoing to develop vaccines against other bacteria. Non-traditional approaches, including phage therapy, microbiome manipulation, virulence factor targeting and immunomodulation are all receiving attention22. A rapid point-of-care diagnostic that could confirm bacterial infections, the type of bacteria, and its resistance profile, would have a dramatic impact of reducing unnecessary antibiotic usage.
Key to the development of new antibiotics are funding mechanisms to advance them through clinical trials. In 2016 the WHO and Drugs for Neglected Diseases initiative (DNDi) founded GARDP, the Global Antibiotic Research and Development Partnership, with the objective of raising EUR 270m to develop a pipeline of pre-clinical and clinical antibiotics. The same year CARB-X (Combating Antibiotic Resistant Bacteria Biopharmaceutical Accelerator) was established by the Wellcome Trust, BARDA (Biomedical Advanced Research and Development Authority), and NIAID (National Institute of Allergy and Infectious Diseases), with subsequent additional partners adding to a pot of over US$500m to bridge the ‘valley of death’ for antibiotics and diagnostics. However, more needs to be done to entice pharmaceutical companies to reinvest and re-engage in antibiotic discovery. DRIVE-AB, an Innovative Medicines Initiative project looking at the economics of antibiotic development, recommended substantial market entry awards of up to $2b23. In January 2019 the United Kingdom Department of Health announced they would explore a new model where companies are paid based on the health-value of their medicines, rather than the amount of antibiotics they sell, to encourage the development of new, high-priority drugs.
Australians are punching well above their weight when it comes to discovering and developing new antibiotics, alternative therapies and diagnostics. A global screening initiative to discover new antibiotics, the Community for Open Antimicrobial Drug Discovery, is based at the University of Queensland (UQ), and has tested nearly 300 000 compounds sent by over 250 collaborating groups from 45 different countries24. Promising new antibiotics have been developed at Monash University25 and UQ25,26, while other groups have discovered compounds that act via novel mechanisms of action, including inhibitors of the key peptidoglycan enzyme glycosyltransferase27, the bacterial sliding clamp involved in DNA replication28, or the essential metabolic enzyme biotin protein ligase (BPL)29. Prana Biotechnology’s (Melbourne) zinc ionophore PBP2 has been repurposed as a ‘resistance breaker’ by UQ researchers30, while Australians are leading investigations into targeting a virulence factor involved in protein disulfide formation31 and in studying efflux pump inhibition to boost antibiotic activity32. Alternative approaches are being pursued by AmpliPhi Biosciences (phage therapy), the Centre for Digestive Diseases (Sydney; microbiome manipulation via fecal transplant therapy), and a UQ probiotic project (for chronic rhinosinusitis)33. Researchers at the University of Western Australia have developed an innovative rapid method to assess antimicrobial resistance profiles using flow cytometry34. Australians are also leading antibiotic clinical trials, both in comparisons of existing therapies (e.g. a trial comparing piperacillin-tazobactam vs meropenem)35 and in first-in-man trials of new antibiotics such as Spero Therapeutics’ oral carbapenem-class antibiotic SPR994 (NCT03395249) and novel resistance breaker SPR741 (NCT03022175), or Achaogen’s β-lactam/lactamase inhibitor ACHN-383/789 (NCT03163550).
In summary, we truly are facing a situation where we could return to life without antibiotics, where people routinely die from simple infections. There must be a concerted effort by academics, industry and government to find new solutions, both to the economic challenges inhibiting antibiotic development, and to the scientific hurdles required to develop new therapeutics.
Conflicts of interest
MATB is an inventor on antibiotic-related patents and consults for several companies developing antimicrobial treatments.
Acknowledgements
This research did not receive any specific funding.
References
[1] D’Costa, V.M. et al. (2011) Antibiotic resistance is ancient. Nature 477, 457–461.| Antibiotic resistance is ancient.Crossref | GoogleScholarGoogle Scholar | 21881561PubMed |
[2] O’Neill, J. (2016) Review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf
[3] Gilbert, J.A. et al. (2018) Current understanding of the human microbiome. Nat. Med. 24, 392–400.
| Current understanding of the human microbiome.Crossref | GoogleScholarGoogle Scholar | 29634682PubMed |
[4] Klein, E.Y. et al. (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 115, E3463–E3470.
| Global increase and geographic convergence in antibiotic consumption between 2000 and 2015.Crossref | GoogleScholarGoogle Scholar | 29581252PubMed |
[5] Van Boeckel, T.P. et al. (2015) Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 112, 5649–5654.
| Global trends in antimicrobial use in food animals.Crossref | GoogleScholarGoogle Scholar | 25792457PubMed |
[6] Fernandes, P. (2015) The global challenge of new classes of antibacterial agents: an industry perspective. Curr. Opin. Pharmacol. 24, 7–11.
| The global challenge of new classes of antibacterial agents: an industry perspective.Crossref | GoogleScholarGoogle Scholar | 26119487PubMed |
[7] Butler, M.S. et al. (2017) Antibiotics in the clinical pipeline at the end of 2015. J. Antibiot. (Tokyo) 70, 3–24.
| Antibiotics in the clinical pipeline at the end of 2015.Crossref | GoogleScholarGoogle Scholar | 27353164PubMed |
[8] Theuretzbacher, U. et al. (2019) Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect. Dis. 19, e40–e50.
| Analysis of the clinical antibacterial and antituberculosis pipeline.Crossref | GoogleScholarGoogle Scholar | 30337260PubMed |
[9] World Health Organization (2017) Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline, including tuberculosis. WHO/EMP/IAU/2017.12, Geneva.
[10] PhRMA (2015) Medicines in development for cancer. http://phrma-docs.phrma.org/sites/default/files/pdf/oncology-report-2015.pdf (accessed 20 May 2018).
[11] von Nussbaum, F. et al. (2006) Antibacterial natural products in medicinal chemistry--exodus or revival? Angewmandte Chemistry International Edition English 45, 5072–5129.
| Antibacterial natural products in medicinal chemistry--exodus or revival?Crossref | GoogleScholarGoogle Scholar |
[12] Payne, D.J. et al. (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40.
| Drugs for bad bugs: confronting the challenges of antibacterial discovery.Crossref | GoogleScholarGoogle Scholar | 17159923PubMed |
[13] Tommasi, R. et al. (2015) ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 14, 529–542.
| ESKAPEing the labyrinth of antibacterial discovery.Crossref | GoogleScholarGoogle Scholar | 26139286PubMed |
[14] Tomaras, A.P. et al. (2014) LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens. MBio 5, e01551-14.
| LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens.Crossref | GoogleScholarGoogle Scholar | 25271285PubMed |
[15] Richter, M.F. et al. (2017) Predictive compound accumulation rules yield a broad – spectrum antibiotic. Nature 545, 299–304.
| Predictive compound accumulation rules yield a broad – spectrum antibiotic.Crossref | GoogleScholarGoogle Scholar | 28489819PubMed |
[16] Ling, L.L. et al. (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459.
| A new antibiotic kills pathogens without detectable resistance.Crossref | GoogleScholarGoogle Scholar | 25561178PubMed |
[17] Clardy, J. et al. (2006) New antibiotics from bacterial natural products. Nat. Biotechnol. 24, 1541–1550.
| New antibiotics from bacterial natural products.Crossref | GoogleScholarGoogle Scholar | 17160060PubMed |
[18] Ueda, K. and Beppu, T. (2017) Antibiotics in microbial coculture. J. Antibiot. (Tokyo) 70, 361–365.
| Antibiotics in microbial coculture.Crossref | GoogleScholarGoogle Scholar | 27756913PubMed |
[19] Khalil, Z.G. et al. (2019) Inter-Kingdom beach warfare: microbial chemical communication activates natural chemical defences. ISME J. 13, 147–158.
| Inter-Kingdom beach warfare: microbial chemical communication activates natural chemical defences.Crossref | GoogleScholarGoogle Scholar | 30116041PubMed |
[20] Machado, H. et al. (2017) Omics-based natural product discovery and the lexicon of genome mining. Curr. Opin. Microbiol. 39, 136–142.
| Omics-based natural product discovery and the lexicon of genome mining.Crossref | GoogleScholarGoogle Scholar | 29175703PubMed |
[21] Hover, B.M. et al. (2018) Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 3, 415–422.
| Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens.Crossref | GoogleScholarGoogle Scholar | 29434326PubMed |
[22] Czaplewski, L. et al. (2016) Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251.
| Alternatives to antibiotics—a pipeline portfolio review.Crossref | GoogleScholarGoogle Scholar | 26795692PubMed |
[23] (2018) Wanted: a reward for antibiotic development. Nature Biotechnology 36, 555.
| Wanted: a reward for antibiotic development.Crossref | GoogleScholarGoogle Scholar | 29979651PubMed |
[24] Blaskovich, M.A.T. et al. (2015) Helping chemists discover new antibiotics. ACS Infect. Dis. 1, 285–287.
| Helping chemists discover new antibiotics.Crossref | GoogleScholarGoogle Scholar |
[25] Velkov, T. et al. (2018) Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-resistant Gram-negative bacteria. Cell Chem. Biol. 25, 380–391.e5.
| Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-resistant Gram-negative bacteria.Crossref | GoogleScholarGoogle Scholar | 29396290PubMed |
[26] Blaskovich, M.A.T. et al. (2018) Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 9, 22.
| Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria.Crossref | GoogleScholarGoogle Scholar |
[27] Zuegg, J. et al. (2015) Carbohydrate scaffolds as glycosyltransferase inhibitors with in vivo antibacterial activity. Nat. Commun. 6, 7719.
| Carbohydrate scaffolds as glycosyltransferase inhibitors with in vivo antibacterial activity.Crossref | GoogleScholarGoogle Scholar | 26194781PubMed |
[28] Yin, Z. et al. (2015) Bacterial sliding clamp inhibitors that mimic the sequential binding mechanism of endogenous linear motifs. J. Med. Chem. 58, 4693–4702.
| Bacterial sliding clamp inhibitors that mimic the sequential binding mechanism of endogenous linear motifs.Crossref | GoogleScholarGoogle Scholar | 25970224PubMed |
[29] Feng, J. et al. (2016) Biotin protein ligase is a target for new antibacterials. Antibiotics (Basel) 5, 26.
| Biotin protein ligase is a target for new antibacterials.Crossref | GoogleScholarGoogle Scholar |
[30] Bohlmann, L. et al. (2018) Chemical synergy between ionophore PBT2 and zinc reverses antibiotic resistance. MBio 9, e02391-18.
| Chemical synergy between ionophore PBT2 and zinc reverses antibiotic resistance.Crossref | GoogleScholarGoogle Scholar | 30538186PubMed |
[31] Heras, B. et al. (2015) Targeting virulence not viability in the search for future antibacterials. Br. J. Clin. Pharmacol. 79, 208–215.
| Targeting virulence not viability in the search for future antibacterials.Crossref | GoogleScholarGoogle Scholar | 24552512PubMed |
[32] Wang, Y. et al. (2018) Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 143, 699–709.
| Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors.Crossref | GoogleScholarGoogle Scholar | 29220791PubMed |
[33] Cervin, A.U. (2018) The potential for topical probiotic treatment of chronic rhinosinusitis, a personal perspective. Front. Cell. Infect. Microbiol. 7, 530.
| The potential for topical probiotic treatment of chronic rhinosinusitis, a personal perspective.Crossref | GoogleScholarGoogle Scholar | 29379772PubMed |
[34] Mulroney, K.T. et al. (2017) Rapid susceptibility profiling of carbapenem-resistant Klebsiella pneumoniae. Sci. Rep. 7, 1903.
| Rapid susceptibility profiling of carbapenem-resistant Klebsiella pneumoniae.Crossref | GoogleScholarGoogle Scholar | 28507322PubMed |
[35] Harris, P.N.A. et al. (2018) Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA 320, 984–994.
| Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial.Crossref | GoogleScholarGoogle Scholar |
Biography
Dr Mark Blaskovich is an antibiotic hunter based at the Centre for Superbug Solutions in the Institute for Molecular Bioscience at The University of Queensland. A medicinal chemist with 15 years of industrial drug development experience prior to his academic career, Mark has been developing new antibiotics to treat drug resistant pathogens and using modified antibiotics to detect bacterial infections. He is a cofounder of the Community for Open Antimicrobial Drug Discovery, a global antibiotic discovery initiative.