Impact of whole genome sequencing in Public Health reference laboratories
Amy V JennisonPublic Health Microbiology, Forensic and Scientific Services
Queensland Department of Health
Coopers Plains
Brisbane, Qld 4108, Australia
Tel: +61 7 3096 2826
Email: Amy.Jennison@health.qld.gov.au
Microbiology Australia 38(4) 168-171 https://doi.org/10.1071/MA17060
Published: 31 October 2017
Public Health Microbiology reference laboratories fulfil a critical role in providing overarching testing and surveillance for notifiable, emerging and important pathogens. These duties require the laboratory to possess an extensive repertoire of validated assays and the ability to rapidly respond to novel threats and outbreaks. For these, among other reasons, the ‘one stop shop’ approach of whole genome sequencing (WGS) has been embraced by microbiology reference laboratories. The ability to replace multiple labour-intensive assays with a single technique of superior typeability and discrimination at an often competitive price, although not without its challenges, has already begun to change the workflow of Public Health reference laboratories.
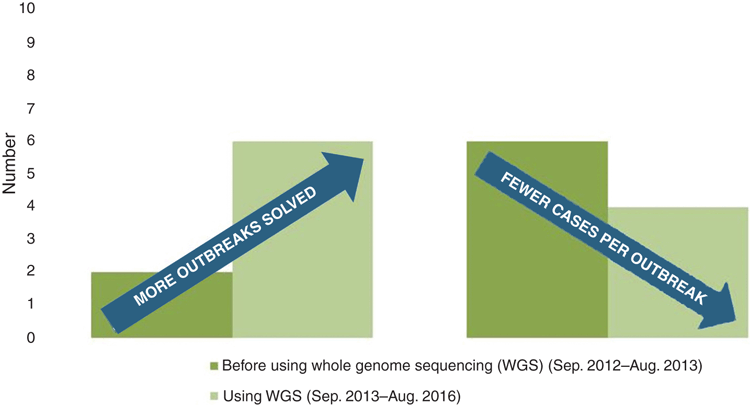
Overseas laboratories such as Public Health England and the US Centers for Disease Control and Prevention (CDC) are exemplars of how WGS has replaced not only conventional molecular typing for the routine surveillance of pathogens but also phenotypic testing such as serotyping1–3. The US Food and Drug Administration (FDA) sequencing initiative, GenomeTrakr consists of a network of US and international laboratories performing real time sequencing on foodborne pathogens from food, environmental and human samples, with sequences being made available on the public NCBI database. This rapid and detailed genomic surveillance has already been highly successful in detecting more outbreaks than prior genotyping whilst restricting the number of cases associated with each outbreak (Figure 1)3. Participation in GenomeTrakr resulted in an Australian listeriosis case with no known cause being linked to a US stone fruit outbreak, demonstrating the importance of integrated international surveillance to monitor what is now a global food chain4.
|
National routine surveillance is beginning to take shape across the Australian State reference laboratories, driven by the Communicable Diseases Genomics Network, of which the reference laboratories belong to. All Australian listeriosis cases are subjected to timely sequencing and national comparison at the Microbiological Diagnostic Unit Public Health Laboratory (MDU PHL) based at The University of Melbourne ensuring outbreaks and disease clusters are detected across state borders5. Recently, the nationally coordinated surveillance of all invasive Neisseria meningitidis infections by genomic sequencing, through a number of state reference laboratories, has commenced. The high-resolution cluster analysis possible from the data has already proven invaluable in understanding the rapid changes to Australian meningococcal disease epidemiology that has occurred over the past two years6.
Public Health Laboratories have also taken the opportunity to replace multiple complex and specialised phenotypic and genotypic tests with the more streamlined process of genomic sequencing. Like other Australian Public Health reference laboratories, the Queensland Public Health Microbiology (PHM) laboratory has moved away from the technical Elek test to testing C. diphtheriae and C. ulcerans by PCR for the diphtheria toxin gene. However, international studies have suggested between 5–10% of PCR positive strains are actually carrying non-functional toxin genes, referred to as non-toxigenic toxin gene bearing (NTTB) isolates. Genetic mutations conferring non-toxigenic status cannot be determined from the screening PCR. By applying WGS, PHM has been able to infer functionality in diphtheria toxin gene positive strains, to not only report the first NTTB isolates from cases tested in Australia but to also characterise the genetic mutations associated with these isolates7. This sequencing is now routinely performed with appropriate timeliness to contribute to the public health response to toxin gene positive cases.
The use of whole genome sequencing to predict antibiotic resistance is still in its infancy. Overall, correlation between the extrapolated genotype and phenotype is promising and the epidemiological information generated on genetic mechanisms is valuable8,9. Reports suggest that inferring antibiotic resistance from genotype results in an overestimation of resistance, possibly due to the presence of silent genes or poorly understood mechanisms9,10. Certainly, there is no doubt that phenotypic testing is still incredibly important and that a greater body of work is necessary and rapid turn-around time established before WGS inferred antimicrobial resistance will be suitable for standalone use in clinical decision-making.
The application of genomics in public health microbiology laboratories is probably best known for its convenience and performance in resolving disease outbreaks and clusters. The ability to respond rapidly, regardless of the organism’s identity, with a technique that offers superior discrimination to conventional typing methods has begun to transform and simplify reference laboratories testing regimes. Public Health Microbiology laboratories are beginning to move away from multi-locus variable number tandem repeat analysis (MLVA), multi-locus sequence typing (MLST) and binary typing based schemes for cluster analysis, instead applying core genome (cgMLST) or whole genome (wgMLST) and single nucleotide polymorphism (SNP)-based typing. The scientific literature contains numerous examples of the retrospective application of WGS to examine outbreaks, which demonstrate the improved discrimination that wgMLST or SNP-based typing offers to either delineating linked cases from sporadic infections or identifying related cases unsuspected through epidemiological data11–13. An increasing number of reports in the literature are also showing that WGS can be utilised to generate real time data able to inform earlier and with increased confidence on ongoing outbreaks3,14. A recent spate of Burkholderia cenocepacia bacteraemia in Australian patients was identified as a point source cluster caused by contaminated ultrasound gel used in central line insertion by whole genome sequencing. The cluster analysis prompted a real time actionable response even as cases were still being identified, including a TGA recall of the product and an international publication calling for healthcare facilities to perform retrospective patient investigations15.
It is also evident that the application of genomics to investigating the transmission of antimicrobial resistant organisms through clinical centres and tracking inter-institutional spread is critical for surveillance and control of resistance. Institutional genomics surveillance programs have demonstrated that WGS based phenotypic tracking can be economically and clinically feasible16,17. Furthermore, WGS has been able to accurately pinpoint not only the ongoing transmission of a resistant strain within the clinical setting but identify the likely point source. An investigation into an MRSA outbreak in a special care baby unit revealed that a baby became infected with the same strain, post deep cleaning and 64 days after the last positive patient. Rapid genomic-based screening revealed a MRSA outbreak strain carrier amongst staff. After relocation and decolonisation therapy of this staff member, no further cases were identified18.
While whole genome sequencing performed on bacterial isolates is well established, metagenomic or deep sequencing directly on clinical specimens has more recently emerged as a valuable technique in public health microbiology. Deep sequencing is particularly useful for the generation of antibiotic resistance data for slow growing or difficult to culture organisms such as Mycobacterium tuberculosis. While PCR based testing is limited in the number of genetic mutations it can target, there is no such limitation for genomic sequencing. A recent paper has reported that they were able to predict antibiotic resistance mechanisms for Mycobacterium tuberculosis by direct deep sequencing on respiratory specimens in less than 48 hours turnaround time, generating results for which standard testing would usually require weeks19.
For other microorganisms, deep sequencing may fill the gap in epidemiological data caused by the increase in culture independent diagnostic testing (CIDT) in pathology laboratories. Molecular diagnosis has many advantages for patient care, however the absence of isolates subsequently available for public health surveillance is concerning. For certain pathogens, such as Neisseria gonorrhoeae, molecular diagnoses can represent up to 80% of disease notifications, meaning resistance and cluster analysis surveillance traditionally only performed on isolates, is fragmented. A recent publication reported success in generating genotyping and antibiotic resistance markers direct from N. gonorrhoeae PCR positive clinical specimens, indicating that epidemiological surveillance is feasible in the absence of culture20.
The use of deep sequencing for direct diagnostics in clinical microbiology is still very much a burgeoning technology, although examples in the literature do showcase the potential21–23. While cost and validity of results are still at times difficult to warrant, the clear advantages of hypothesis-free testing with no prior knowledge of the causative agent required or issues around mutational changes in primer regions offers tantalising prospects to the clinical microbiology field24. As sequencing costs decrease and long read technologies become more accessible, it is likely that current issues around financial justification, sensitivity, validation and clinical interpretation will be addressed.
Despite the many advantages WGS brings to public health microbiology, the integration into laboratories does not come without a suite of challenges. Traditionally wet lab-based laboratories must become au fait with high power computing infrastructure, generate solutions for the handling and storage of big data, acquire bioinformatics skills and establish interpretation and management of complex data. Laboratory staff must not only become familiar with these new analyses and solve reporting and LIMS challenges around the use of constantly changing dendrograms and SNP differences, but must engage in the re-education of clinicians, government bodies and public health officers. Accreditation to ensure robustness of WGS analysis is essential but is still in early phases for most laboratories and with the respective regulatory bodies, acceptance criteria and analysis still far from standardised at a national, let alone international level, and to date only a handful of international QAPs are available. It is essential that accreditation is appropriate and handled by subject matter experts as it is not amenable to simply apply established human clinical genetics requirements to public health microbiology genomics. While standardisation of SNP typing is still complex and requires centralised analysis, wgMLST with the development of online, curated, freely accessible databases may fill the specifications for stable standardised analogous analysis for at least some bacterial species.
The current challenges, however, are far from untenable and it is clear that as the cost continues to decrease, so that replacement of more standard microbiology tests becomes feasible, and standardisation efforts at both the national and international level progress, that WGS will only continue to revolutionise testing strategies employed in public health microbiology.
References
[1] Ashton, P.M. et al. (2016) Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4, e1752.| Identification of Salmonella for public health surveillance using whole genome sequencing.Crossref | GoogleScholarGoogle Scholar |
[2] Chattaway, M.A. et al. (2016) Whole genome sequencing for public health surveillance of Shiga toxin-producing Escherichia coli other than serogroup O157. Front. Microbiol. 7, 258.
| Whole genome sequencing for public health surveillance of Shiga toxin-producing Escherichia coli other than serogroup O157.Crossref | GoogleScholarGoogle Scholar |
[3] Jackson, B.R. et al. (2016) Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin. Infect. Dis. 63, 380–386.
| Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation.Crossref | GoogleScholarGoogle Scholar |
[4] Kwong, J.C. et al. (2016) Sharing is caring: international sharing of data enhances genomic surveillance of Listeria monocytogenes. Clin. Infect. Dis. 63, 846–848.
| Sharing is caring: international sharing of data enhances genomic surveillance of Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar |
[5] Kwong, J.C. et al. (2016) Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 54, 333–342.
| Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xps1ensLs%3D&md5=eedd228d2a7d589510de704de34b1417CAS |
[6] Martin, N.V. et al. (2016) Rise in invasive serogroup W meningococcal disease in Australia 2013–2015. Commun. Dis. Intell. Q. Rep. 40, E454–E459.
[7] Doyle, C.J. et al. (2017) Sequence analysis of toxin gene-bearing Corynebacterium diphtheriae strains, Australia. Emerg. Infect. Dis. 23, 105–107.
| Sequence analysis of toxin gene-bearing Corynebacterium diphtheriae strains, Australia.Crossref | GoogleScholarGoogle Scholar |
[8] Ellington, M.J. et al. (2017) The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 23, 2–22.
| The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC2sjgslChsQ%3D%3D&md5=f43f996f2c792ec5b792efcdc17c41c2CAS |
[9] McDermott, P.F. et al. (2016) Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60, 5515–5520.
| Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhtlKksLg%3D&md5=ae4eb4fcea89c0de0c3a7c684f1b5e99CAS |
[10] Hasman, H. et al. (2014) Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146.
| Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples.Crossref | GoogleScholarGoogle Scholar |
[11] Kovanen, S.M. et al. (2014) Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland. J. Clin. Microbiol. 52, 4147–4154.
| Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland.Crossref | GoogleScholarGoogle Scholar |
[12] Octavia, S. et al. (2015) Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. J. Clin. Microbiol. 53, 1063–1071.
| Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXlsFSlt7Y%3D&md5=2a60292c1f852afb28ad4badf5dddca4CAS |
[13] Ruppitsch, W. et al. (2015) Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 53, 2869–2876.
| Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xpt1Oqsbg%3D&md5=56d93c4bc3d760bdd464baae1ba9c8bfCAS |
[14] McGann, P. et al. (2016) Real time application of whole genome sequencing for outbreak investigation – what is an achievable turnaround time? Diagn. Microbiol. Infect. Dis. 85, 277–282.
| Real time application of whole genome sequencing for outbreak investigation – what is an achievable turnaround time?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xns1egurk%3D&md5=b8961fc45a4c0027c52a109a647428eaCAS |
[15] Shaban, R.Z. et al. (2017) Outbreak of health care-associated Burkholderia cenocepacia bacteremia and infection attributed to contaminated sterile gel used for central line insertion under ultrasound guidance and other procedures. Am. J. Infect. Control 45, 954–958.
| Outbreak of health care-associated Burkholderia cenocepacia bacteremia and infection attributed to contaminated sterile gel used for central line insertion under ultrasound guidance and other procedures.Crossref | GoogleScholarGoogle Scholar |
[16] Gorrie, C.L. et al. (2017) Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215.
| Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients.Crossref | GoogleScholarGoogle Scholar |
[17] Mellmann, A. et al. (2016) Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J. Clin. Microbiol. 54, 2874–2881.
| Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting.Crossref | GoogleScholarGoogle Scholar |
[18] Harris, S.R. et al. (2013) Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 13, 130–136.
| Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1Gksr0%3D&md5=d472aea2ba49b3ff81961c7e20b0df84CAS |
[19] Votintseva, A.A. et al. (2017) Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 55, 1285–1298.
| Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples.Crossref | GoogleScholarGoogle Scholar |
[20] Graham, R.M. et al. (2017) Epidemiological typing of Neisseria gonorrhoeae and detection of markers associated with antimicrobial resistance directly from urine samples using next generation sequencing. Sex. Transm. Infect. 93, 65–67.
| Epidemiological typing of Neisseria gonorrhoeae and detection of markers associated with antimicrobial resistance directly from urine samples using next generation sequencing.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC28jpsV2jtA%3D%3D&md5=1947e2550ce16435aa541074904c708bCAS |
[21] Kuroda, M. et al. (2012) Detection of a possible bioterrorism agent, Francisella sp., in a clinical specimen by use of next-generation direct DNA sequencing. J. Clin. Microbiol. 50, 1810–1812.
| Detection of a possible bioterrorism agent, Francisella sp., in a clinical specimen by use of next-generation direct DNA sequencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XnsF2ksrc%3D&md5=fa39b0e365076b7645ae32d8ad1fbfabCAS |
[22] Salzberg, S.L. et al. (2016) Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol. Neuroimmunol. Neuroinflamm. 3, e251.
| Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system.Crossref | GoogleScholarGoogle Scholar |
[23] Wilson, M.R. et al. (2014) Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 370, 2408–2417.
| Actionable diagnosis of neuroleptospirosis by next-generation sequencing.Crossref | GoogleScholarGoogle Scholar |
[24] Allcock, R.J.N. et al. (2017) Towards a universal molecular microbiological test. J. Clin. Microbiol. , .
| Towards a universal molecular microbiological test.Crossref | GoogleScholarGoogle Scholar |
Biography
Dr Amy Jennison is the Supervising Scientist of Molecular Epidemiology, Public Health Microbiology, which is the Queensland reference laboratory responsible for the molecular surveillance of notifiable bacterial pathogens and characterisation of public health related outbreaks. She is leading a team in the application of WGS to routine molecular surveillance and heads numerous research projects aimed at utilising WGS for improving molecular epidemiological investigation and addressing culture independent diagnostic testing through deep sequencing approaches.


