The laboratory diagnosis of Strongyloides stercoralis
Matthew R Watts A * , Gemma Robertson B * and Richard S Bradbury C DA Centre for Infectious Diseases and Microbiology, Pathology West – ICMPR, and Marie Bashir Institute, University of Sydney, Westmead Hospital, Westmead, Sydney, NSW, Australia, Tel: +61 2 9845 6255, Email: matthew.watts@health.nsw.gov.au
B Melbourne Pathology, Collingwood and James Cook University, Tel: +61 3 9287 7700, Email: gemmajrobertson@gmail.com
C School of Medical and Applied Sciences, Central Queensland University, Rockhampton, Qld, Australia
D Corresponding author. Email: r.bradbury@cqu.edu.au
Microbiology Australia 37(1) 4-9 https://doi.org/10.1071/MA16003
Published: 12 February 2016
It is estimated that over 30 million people worldwide are infected by the nematode, Strongyloides stercoralis1. It is endemic in sub-tropical and tropical parts of Australia, with high rates of infection documented in some indigenous communities2. Due to the potential for chronic autoinfection, that may persist for decades, migration leads to the presence of the infection in non-endemic areas1. Transmission to humans is generally through the penetration of larvae through the skin, following contact with faecally contaminated soil1. Disease severity ranges from asymptomatic chronic carriage to an overwhelming illness, where large numbers spread throughout the body, usually triggered by immunosuppression1.
Clinicians are advised to consider strongyloidiasis in patients prior to immunosuppression, or with indicative symptoms, if there is a history of probable exposure in an endemic area, regardless of the elapsed time since exposure3,4. Eosinophilia is not an accurate marker of strongyloidiasis, with a retrospective study finding that only a quarter of patients with Strongyloides infection had a raised eosinophil count5. The detection of strongyloidiasis is optimised by appropriate test ordering, clinical notes, specimen transportation, and processing by the receiving laboratory.
The gold standard for the diagnosis of strongyloidiasis is the morphological identification of larvae in stool, tissue biopsies, and other clinical specimens such as bronchoalveolar lavage. However, in chronic infections, detection can be limited by low larval output in stool, leading to false negative results6. Consequently, in validation studies for serological and nucleic acid tests there is a tendency to define heavier infections as ‘true positives’. This affects serological cut-offs, measurements of sensitivity and specificity, and positive and negative predictive values7. Recognition of these limitations is important for the interpretation of negative diagnostic test results, where clinical suspicion remains. Here, we will give an overview of currently available conventional and molecular tests for the diagnosis of strongyloidiasis.
Stool microscopy and culture methods
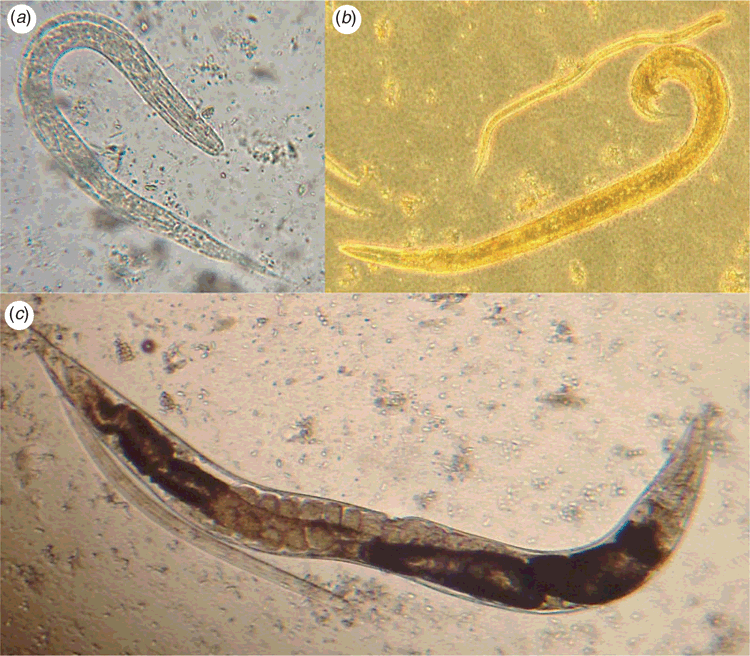
Specimen transport and storage have a major impact on the efficacy of culture techniques in the laboratory diagnosis of S. stercoralis from faecal samples. Fresh, unrefrigerated samples should be delivered to the laboratory for culture as soon after collection as possible, as the viability of larvae decreases incrementally with storage at 4°C over a 72-h period8. Rhabditiform and filariform larvae will be found along with free-living adults of S. stercoralis in older cultures (Figure 1). Larval stages must be differentiated from those of hookworms, which may also be recovered.
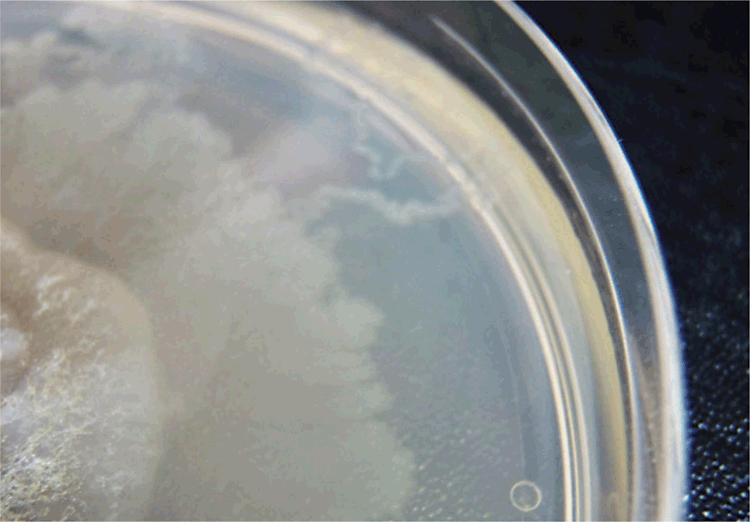
Microscopic methodologies such as examination of Kato-Katz preparations, FLOTAC, and formalin/ethyl acetate concentrates have a low yield compared to culture9,10. A modified formalin/ethyl acetate method proposed by Anamnart et al. improved rates of detection9. Overall, however, microscopic techniques alone are insensitive and not sufficient for the exclusion of strongyloidiasis. In one of these studies, though 30 of 254 participants were diagnosed with strongyloidiasis by either agar plate culture (APC; Figure 2) or Baermann culture techniques, no infections were identified by microscopy using the Kato-Katz technique11.
APC is possibly the easiest culture to perform in the context of high volume diagnostic testing. Results are available within two days, although extended incubation up to four days increases yield8,11,12. Two studies comparing 48 h APC with Baermann culture found an improved recovery of S. stercoralis larvae in APC11,12. Recovery rates improve markedly with multiple stool cultures6,11,13
Serological diagnosis
Several tests for the serological diagnosis of strongyloidiasis have been described, using both crude and recombinant antigens. Two commercial ELISA kits employing somatic antigens are available from BORDIER (Strongyloides ratti antigen) and IVD Research (S. stercoralis antigen), respectively14. Recently, two recombinant antigens (32 kD recombinant antigen, called NIE and S. stercoralis immmunoreactive antigen, SsIR) have been employed for serological testing in both ELISA and luciferase immunoprecipitation system assay (LIPS) platforms15. The reported sensitivity and specificity of various serological platforms ranges from 56-100% and 29-100%, dependent upon the method, antigens, cut-offs, study populations, and reference methods employed16. Strongyloides serology using a crude larval extract antigen was shown in one study to be less sensitive for the diagnosis of returned travellers (73%) compared to patients who have lived for an extended period in an endemic area (98%)17. No definitive study of serological methods has been conducted to date, and much of the available data is subject to flaws in methodology, particularly the use of microscopy only as a reference standard for positive specimens and varying S. stercoralis exposure rates amongst tested serum groups16. A 2010 study with a reference standard of a combination of three culture methods and sedimentation concentration found NIE LIPS had a sensitivity of 97.8% (cut-off 37.89 LU) in a study population with high endemicity but from regions without filarial infection15. Lower sensitivity resulted when testing the same samples by NIE ELISA(84%), NIE-SsIR LIPS (91.2) and a S. stercoralis crude antigen extract ELISA (97%)18. All assays tested showed 100% specificity in this study15. A more recent study compared an in-house crude S. stercoralis filariform larvae immunofluorescent antibody test (IFAT) with the ELISAs from IVD Research, Bordier, and a recombinant antigen NIE ELISA and LIPS14. This study used reference samples identified as positive by culture as well as microscopy, and also a composite reference standard of concordant results in at least three of five serological tests14. The in-house IFAT was found to be the most sensitive (93.9%) when used in a test subject group with no known previous exposure to S. stercoralis and using the composite reference standard, whilst NIE LIPS was found to be the most specific test (100%)14. Furthermore, when tested against subjects with potential previous exposure and using the composite reference standard, NIE LIPS was almost 100% specific and 84.6% sensitive (cut off value 1388 LU)14. In testing against the same sample group, the Bordier and IVD ELISAs maintained a high specificity (almost 100%), but a lower sensitivity (70% and 79%, respectively) and the NIE ELISA showed the highest specificity (99%), but a low sensitivity (45%) (cut-off 76.5 U/mL)14.
Seroreversion following treatment of many, but not all, patients was noted in a study using a Strongyloides ratti antigen ELISA3. However, this effect is not universal and varies between studies3,17. Immunosuppression was demonstrated to cause a reduction in serological sensitivity (62% vs previously determined 92%), when testing haematological patients on antineoplastic therapies18. NIE LIPS did not cross-react with antigens from other parasites in the study by Bisoffi et al., whereas IFAT and the two commercial ELISAs did yield false positives, particularly from Mansonella perstans infection14. Such cross-reaction may be decreased by pre-incubation of serum in an extract of Onchocerca gutterosa16.
Nucleic acid tests
Nucleic acid tests complement non-molecular methodologies for the diagnosis of S. stercoralis, and allow the use of refrigerated, frozen, or preserved specimens11,19,20. This simplifies specimen transportation, particularly where collection occurs some distance from the testing laboratory, and there is no risk of laboratory-acquired strongyloidiasis21. DNA extraction and amplification can be performed within 1 day, however, laboratories may batch specimens according to demand.
DNA extraction
It is important that DNA extraction methods for stool specimens are effective at removing the numerous nucleic acid test inhibitors in stool22. A comparison of 5 methods of DNA extraction demonstrated that two column-based methods were the most effective for the PCR detection of DNA from Strongyloides ratti that had been spiked into human stool. These were the MoBio PowerSoil kit (MoBio Laboratories, Carlsbad, CA, USA) and a method based on modifications of the QiaAmp Tissue kit (Qiagen, Hilden, Germany) by Verweij et al., which has been successfully automated23. The comparison found that bead beating prior to the use of the NucliSens EasyMag (BioMerieux, Marcy l’Etoile, France) was less effective, which indicates the method of sample pretreatment prior to automated extraction will impact upon test sensitivity. Other investigators have used a variety of different extraction methods for Strongyloides PCR, including in-house methods, the Qiagen stool kit (unmodified and modified), and the Nucleospin Soil kit (Macherey-Nagel, Duren, Germany)10,24–30.
One of the inherent limitations of the molecular diagnosis of S. stercoralis is the sampling error that can occur when relatively small amounts are extracted in the context of low larval output6. For example, 2g of stool can be used for agar plate culture, whereas 250 mg of specimen is recommended for the MoBio PowerSoil kit31. Methods that concentrate larger amounts of stool prior to DNA extraction have the potential to increase test sensitivity, if they remove inhibitors and retain larvae29.
PCR
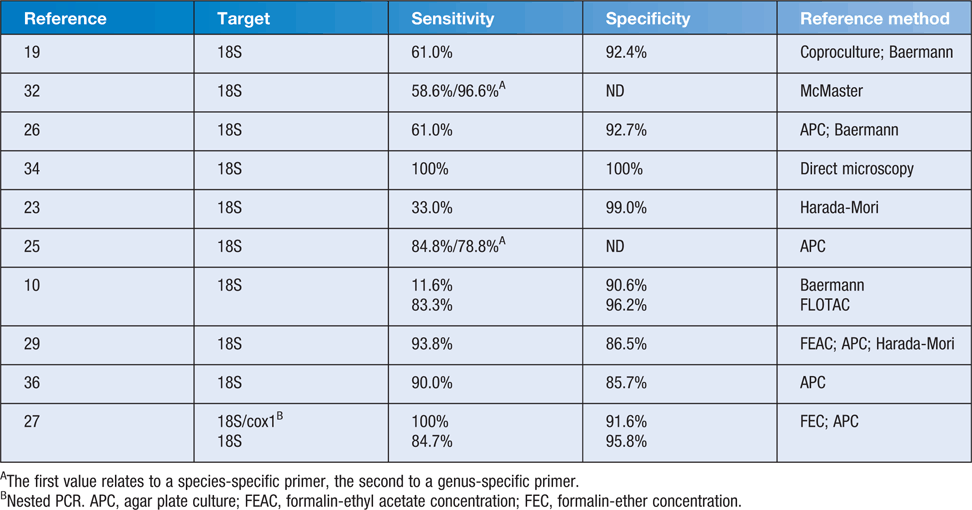
Current PCR methods most commonly target one of four regions: the 18S rRNA small subunit (SSU); the internal transcribed spacer region 1 (ITS-1); the 28S rRNA gene; or the cyclooxygenase gene (cox1)19,23–30,32–37. Published sensitivities and specificities for Strongyloides PCR vary according to the reference methods and are listed in Table 1. The majority of Strongyloides PCR publications have used a real-time method with primers and probe published by Verweij et al.19. This has also allowed for the development of multiplexed PCR10,30,34,38. Some studies evaluating the diagnostic accuracy of these PCR methods have used both morphological diagnosis and detection of PCR products as their reference standards, and are not reviewed here. Their methodology precludes the calculation of sensitivity and specificity based on gold-standard, according to an FDA Guidance39.
|
In the absence of a consistent gold standard in chronic infection, positive nucleic acid test results, where conventional tests are negative, may be due to greater sensitivity or false positive results6. No PCR studies have reported false positive results when analytical specificity has been tested using DNA extracted from bacteria, viruses, fungi, protozoa, and other helminths19,23,24,27,29,30. Studies have also assessed the specificity of the PCR products by sequence analysis, with all finding 100% sequence homology with the target sequence of S. stercoralis.24,25,27,29. Sitta et al. found a number of false positives, using published genus and species-specific primers, based on non-target sized bands on gel electrophoresis19,25. The genus-specific primers amplified sequences that generated non-target bands on electrophoresis in specimens that contained Blastocytis and other helminths on microscopy, and the species-specific primers amplified sequences that generated non-target bands on electrophoresis in specimens positive for hookworm on microscopy25. Similar accounts of cross-reactivity have not yet been reported, so further data will be useful to monitor the specificity of PCR in different populations.
LAMP
Loop-mediated isothermal amplification (LAMP) is an additional nucleic acid detection method. LAMP uses a DNA polymerase with strand-displacement activity, so it doesn’t require the temperature cycling of PCR, and can be performed with a simple source of constant temperature such as a heating block40. LAMP has been successfully applied in resource limited-settings for the detection of pathogens40.
The Strongyloides LAMP assay uses primers that are genus specific and bind to the 28S rRNA gene20. The reaction runs at 60°C for 1 hour. Pre-heating of the reagents and DNA template to 95°C, prior to the addition of enzyme, increases the limit of detection and eliminates the need to pre-heat the template and keep it at 4°C20. A novel use of Syto-82 dye (Life Technologies, Carlsbad, CA, USA) enables the detection of positive results in real-time or visually on completion of the reaction20. Analytical sensitivity and specificity are comparable to PCR, according to the method of Verweij et al.19,20,23. When 28 human stool specimens that were microscopy and PCR positive for S. stercoralis were tested with the LAMP method, 27 were positive20. The negative specimen had a high cycle threshold (38.44) on PCR20. Further validation of the LAMP assay with clinical specimens is currently in progress.
Conclusion
The diagnosis of strongyloidiasis can be made through the morphological identification of larvae, usually in the stool, serological testing, and nucleic acid tests. While each methodology has advantages, there are limitations that need to be taken into account when assessing the significance of negative test results. Often the most important aspect of patient management is to consider the possibility of S. stercoralis infection.
References
[1] Olsen, A. et al. (2009) Strongyloidiasis – the most neglected of the neglected tropical diseases? Trans. R. Soc. Trop. Med. Hyg. 103, 967–972.| Strongyloidiasis – the most neglected of the neglected tropical diseases?Crossref | GoogleScholarGoogle Scholar | 19328508PubMed |
[2] Adams, M. et al. (2003) Strongyloidiasis: an issue in Aboriginal communities. Rural Remote Health 3, 152.
| 1:STN:280:DC%2BD2M3kt1SmsA%3D%3D&md5=a788a19b1ec1aab8b1f64cadedb5d9a2CAS | 15877491PubMed |
[3] Page, W.A. et al. (2006) Utility of serological follow-up of chronic strongyloidiasis after antihelminthic chemotherapy. Trans. R. Soc. Trop. Med. Hyg. 100, 1056–1062.
| Utility of serological follow-up of chronic strongyloidiasis after antihelminthic chemotherapy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFOjsr3M&md5=90bc8adb35622d6d1b84e8fc32ecc543CAS | 16551471PubMed |
[4] Bailey, M.S. et al. (2006) Helminth infections in British troops following an operation in Sierra Leone. Trans. R. Soc. Trop. Med. Hyg. 100, 842–846.
| Helminth infections in British troops following an operation in Sierra Leone.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD28vgvFSruw%3D%3D&md5=e4b573000839eb2466f6d57ffcd2c4faCAS | 16406097PubMed |
[5] Naidu, P et al. (2013) Eosinophilia: a poor predictor of Strongyloides infection in refugees. Can. J. Infect. Dis Med. Microbiol. 24, 93–96.
| 24421809PubMed |
[6] Dreyer, G. et al. (1996) Patterns of detection of Strongyloides stercoralis in stool specimens: implications for diagnosis and clinical trials. J. Clin. Microbiol. 34, 2569–2571.
| 1:STN:280:DyaK2s%2FjsVyltw%3D%3D&md5=c51ee123e2225e93bf4d19579208b63aCAS | 8880521PubMed |
[7] Siddiqui, A.A. and Berk, S.L. (2001) Diagnosis of Strongyloidies stercoralis infection. Clin. Infect. Dis. 33, 1040–1047.
| Diagnosis of Strongyloidies stercoralis infection.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3MvosFGksw%3D%3D&md5=14e38b9087b3990d4ceb5c04429b3eaeCAS | 11528578PubMed |
[8] Inês, EdeJ. et al. (2011) Efficacy of parasitological methods for the diagnosis of Strongyloides stercoralis and hookworm in faecal specimens. Acta Trop. 120, 206–210.
| Efficacy of parasitological methods for the diagnosis of Strongyloides stercoralis and hookworm in faecal specimens.Crossref | GoogleScholarGoogle Scholar |
[9] Anamnart, W. et al. (2010) Factors affecting the recovery of Strongyloides stercoralis larvae: an approach to a newly modified formalin-ether concentrationtechnique for diagnosis of strongyloidiasis. J. Clin. Microbiol. 48, 97–100.
| Factors affecting the recovery of Strongyloides stercoralis larvae: an approach to a newly modified formalin-ether concentrationtechnique for diagnosis of strongyloidiasis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhvFKqtbo%3D&md5=5e6a2891215012f58d91fb6299883751CAS | 19923489PubMed |
[10] Knopp, S. et al. (2014) Diagnostic accuracy of Kato-Katz, FLOTAC, Baermann, and PCR methods for the detection of light-intensity hookworm and Strongyloides stercoralis infections in Tanzania. Am. J. Trop. Med. Hyg. 90, 535–545.
| Diagnostic accuracy of Kato-Katz, FLOTAC, Baermann, and PCR methods for the detection of light-intensity hookworm and Strongyloides stercoralis infections in Tanzania.Crossref | GoogleScholarGoogle Scholar | 24445211PubMed |
[11] Steinmann, P. et al. (2007) Occurrence of Strongyloides stercoralis in Yunnan Province, China, and comparison of diagnostic methods. PLoS Negl. Trop. Dis. 1, e75.
| Occurrence of Strongyloides stercoralis in Yunnan Province, China, and comparison of diagnostic methods.Crossref | GoogleScholarGoogle Scholar | 17989788PubMed |
[12] Khieu, V. et al. (2013) Diagnosis, treatment and risk factors of Strongyloides stercoralis in schoolchildren in Cambodia. PLoS Negl. Trop. Dis. 7, e2035.
| Diagnosis, treatment and risk factors of Strongyloides stercoralis in schoolchildren in Cambodia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXktlSkt74%3D&md5=1ed9ecee63b2e04670cf1fec2dd48514CAS | 23409200PubMed |
[13] Hirata, T. et al. (2007) Short report: increased detection rate of Strongyloides stercoralis by repeated stool examinations using the agar plate culture method. Am. J. Trop. Med. Hyg. 77, 683–684.
| 17978071PubMed |
[14] Bisoffi, Z. et al. (2014) Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection. PLoS Negl. Trop. Dis. 8, e2640.
| Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection.Crossref | GoogleScholarGoogle Scholar | 24427320PubMed |
[15] Krolewiecki, A.J. et al. (2010) Improved diagnosis of Strongyloides stercoralis using recombinant antigen-based serologies in a community-wide study in northern Argentina. Clin. Vaccine Immunol. 17, 1624–1630.
| Improved diagnosis of Strongyloides stercoralis using recombinant antigen-based serologies in a community-wide study in northern Argentina.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXht1Kisr%2FN&md5=f7b5bd61be209fdb2585dcedfcdf3a1aCAS | 20739501PubMed |
[16] Requena-Méndez, A, Chiodini, P, Bisoffi, Z, Buonfrate, D, Gotuzzo, E and Munoz, J (2013) The laboratory diagnosis and follow up of strongyloidiasis: a systematic review. PLoS Negl Trop Dis. 7, e2002.
| 23350004PubMed |
[17] Sudarshi, S. et al. (2003) Clinical presentation and diagnostic sensitivity of laboratory tests for Strongyloides stercoralis in travellers compared with immigrants in a non-endemic country. Trop. Med. Int. Health 8, 728–732.
| Clinical presentation and diagnostic sensitivity of laboratory tests for Strongyloides stercoralis in travellers compared with immigrants in a non-endemic country.Crossref | GoogleScholarGoogle Scholar | 12869094PubMed |
[18] Schaffel, R. et al. (2001) The value of an immunoenzymatic test (enzyme-linked immunosorbent assay) for the diagnosis of strongyloidiasis in patients immunosuppressed by haematologic malignancies. Am. J. Trop. Med. Hyg. 65, 346–350.
| 1:STN:280:DC%2BD3MnjvVeqtQ%3D%3D&md5=7238d98cd355d20f141dfac01f61976eCAS | 11693882PubMed |
[19] Verweij, J.J. et al. (2009) Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans. R. Soc. Trop. Med. Hyg. 103, 342–346.
| Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnsF2jt7k%3D&md5=1f3094517b1da9c884291f6d9edfd63bCAS | 19195671PubMed |
[20] Watts, M.R. et al. (2014) A loop-mediated isothermal amplification (LAMP) assay for Strongyloides stercoralis in stool that uses a visual detection method with SYTO-82 fluorescent dye. Am. J. Trop. Med. Hyg. 90, 306–311.
| A loop-mediated isothermal amplification (LAMP) assay for Strongyloides stercoralis in stool that uses a visual detection method with SYTO-82 fluorescent dye.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXltlyksLY%3D&md5=9eb2e611a2f96e99730a33a974c73919CAS | 24323513PubMed |
[21] Herwaldt, B.L. (2001) Laboratory-acquired parasitic infections from accidental exposures. Clin. Microbiol. Rev. 14, 659–688.
| Laboratory-acquired parasitic infections from accidental exposures.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3MrjvVynug%3D%3D&md5=a01e3b423ac6af4e5fc7932c6c5743b8CAS | 11585780PubMed |
[22] Schrader, C. et al. (2012) PCR inhibitors – occurrence, properties and removal. J. Appl. Microbiol. 113, 1014–1026.
| PCR inhibitors – occurrence, properties and removal.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsFWisrjK&md5=9d700a5d74d20b2dfde402a7478052eeCAS | 22747964PubMed |
[23] Sultana, Y. et al. (2013) Real-time polymerase chain reaction for detection of Strongyloides stercoralis in stool. Am. J. Trop. Med. Hyg. 88, 1048–1051.
| Real-time polymerase chain reaction for detection of Strongyloides stercoralis in stool.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXpvFaqu78%3D&md5=5115f470ea48ec8ead3342faee52af70CAS | 23568289PubMed |
[24] Repetto, S.A. et al. (2013) An improved DNA isolation technique for PCR detection of Strongyloides stercoralis in stool samples. Acta Trop. 126, 110–114.
| An improved DNA isolation technique for PCR detection of Strongyloides stercoralis in stool samples.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXksFCjs7k%3D&md5=262a959a84e0b6c1c123fa0d4ef38825CAS | 23416126PubMed |
[25] Sitta, R.B. et al. (2014) Conventional PCR for molecular diagnosis of human strongyloidiasis. Parasitology 141, 716–721.
| Conventional PCR for molecular diagnosis of human strongyloidiasis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXntFOnu70%3D&md5=a16a590bf8c83915903d23f7504c8c0bCAS | 24476900PubMed |
[26] Schär, F. et al. (2013) Evaluation of real-time PCR for Strongyloides stercoralis and hookworm as diagnostic tool in asymptomatic schoolchildren in Cambodia. Acta Trop. 126, 89–92.
| Evaluation of real-time PCR for Strongyloides stercoralis and hookworm as diagnostic tool in asymptomatic schoolchildren in Cambodia.Crossref | GoogleScholarGoogle Scholar | 23298731PubMed |
[27] Sharifdini, M. et al. (2015) Comparison of nested polymerase chain reaction and real-time polymerase chain reaction with parasitological methods for detection of Strongyloides stercoralis in human fecal samples. Am. J. Trop. Med. Hyg. 93, 1285–1291.
| 26350449PubMed |
[28] Angal, L. et al. (2015) Determining intestinal parasitic infections (IPIs) in inmates from Kajang Prison, Selangor, Malaysia for improved prison management. BMC Infect. Dis. 15, 467.
| Determining intestinal parasitic infections (IPIs) in inmates from Kajang Prison, Selangor, Malaysia for improved prison management.Crossref | GoogleScholarGoogle Scholar | 26511347PubMed |
[29] Saugar, J.M. et al. (2015) Application of real-time PCR for the detection of Strongyloides spp. in clinical samples in a reference center in Spain. Acta Trop. 142, 20–25.
| Application of real-time PCR for the detection of Strongyloides spp. in clinical samples in a reference center in Spain.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvFSms73I&md5=9e403db10f3a94971b1f144ced7a3304CAS | 25447829PubMed |
[30] Janwan, P. et al. (2011) Rapid detection of Opisthorchis viverrini and Strongyloides stercoralis in human fecal samples using a duplex real-time PCR and melting curve analysis. Parasitol. Res. 109, 1593–1601.
| Rapid detection of Opisthorchis viverrini and Strongyloides stercoralis in human fecal samples using a duplex real-time PCR and melting curve analysis.Crossref | GoogleScholarGoogle Scholar | 21537984PubMed |
[31] Koga, K. et al. (1991) A modified agar plate method for detection of Strongyloides stercoralis. Am. J. Trop. Med. Hyg. 45, 518–521.
| 1:STN:280:DyaK38%2FlvFKksA%3D%3D&md5=c5fd47ffdf22ef211a9d999305c82087CAS | 1951861PubMed |
[32] Marra, N.M. et al. (2010) Faecal examination and PCR to detect Strongyloides venezuelensis in experimentally infected Lewis rats. Mem. Inst. Oswaldo Cruz 105, 57–61.
| Faecal examination and PCR to detect Strongyloides venezuelensis in experimentally infected Lewis rats.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXntFyiurg%3D&md5=eb68429e292a8b988ab745c11a5cb762CAS | 20209330PubMed |
[33] Basuni, M. et al. (2011) A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am. J. Trop. Med. Hyg. 84, 338–343.
| A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths.Crossref | GoogleScholarGoogle Scholar | 21292911PubMed |
[34] Mejia, R. et al. (2013) A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am. J. Trop. Med. Hyg. 88, 1041–1047.
| A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXpvFaqurc%3D&md5=ace73f51bf9efd78bdaf70508d2da6e8CAS | 23509117PubMed |
[35] Zueter, A.M. et al. (2014) Detection of Strongyloides stercoralis infection among cancer patients in a major hospital in Kelantan, Malaysia. Singapore Med. J. 55, 367–371.
| Detection of Strongyloides stercoralis infection among cancer patients in a major hospital in Kelantan, Malaysia.Crossref | GoogleScholarGoogle Scholar | 25091885PubMed |
[36] de Paula, F.M. et al. (2015) Molecular diagnosis of strongyloidiasis in tropical areas: a comparison of conventional and real-time polymerase chain reaction with parasitological methods. Mem. Inst. Oswaldo Cruz 110, 272–274.
| Molecular diagnosis of strongyloidiasis in tropical areas: a comparison of conventional and real-time polymerase chain reaction with parasitological methods.Crossref | GoogleScholarGoogle Scholar |
[37] Moghaddassani, H. et al. (2011) Molecular diagnosis of Strongyloides stercoralis infection by PCR detection of specific DNA in human stool samples. Iran. J. Parasitol. 6, 23–30.
| 1:CAS:528:DC%2BC3MXht1equr7K&md5=86cb850129f80d7785cf9849c477202eCAS | 22347284PubMed |
[38] Basuni, M. et al. (2012) Detection of selected intestinal helminths and protozoa at Hospital Universiti Sains Malaysia using multiplex real-time PCR. Trop. Biomed. 29, 434–442.
| 1:STN:280:DC%2BC3s%2FgslKjsg%3D%3D&md5=c075aa1282fc31007565bc69c58db819CAS | 23018507PubMed |
[39] U.S. Food and Drug Administration (2007) Statistical guidance on reporting results from studies evaluating diagnostic tests. http://www.fda.gov/RegulatoryInformation/Guidances/ucm071148.htm.
[40] Mori, Y. et al. (2013) Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J. Infect. Chemother. 19, 404–411.
| Loop-mediated isothermal amplification (LAMP): recent progress in research and development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXps1yht74%3D&md5=df35e8ec535dea3940f873697ea88745CAS | 23539453PubMed |
* These two authors contributed equally.
Biographies
Matthew Watts is an Infectious Diseases Physician and Clinical Microbiologist based at the Centre for Infectious Diseases and Microbiology, Pathology West-ICPMR and the Marie Bashir Institute, University of Sydney, Westmead Hospital. His interests include parasitic and zoonotic infections.
Gemma Robertson is a final year microbiology trainee at Melbourne Pathology. She has an interest in tropical medicine and parasitology, and will be undertaking a PhD to continue her research into soil-transmitted helminthiases in Aboriginal communities.
Dr Richard Bradbury is an Australian Parasitologist with an interest in all fields of parasitology. He was recently appointed as the Team Lead in the Parasite Diagnostics and Biology Laboratory of the Centers for Disease Control and Prevention in Atlanta, USA. He is writing this work in both his personal capacity and in his capacity as an adjunct academic at Central Queensland University.