You are what you secrete: extracellular proteins and virulence in Cryptococcus
Leona T Campbell A , Matthew P Padula B , Elizabeth Harry B and Dee A Carter A CA School of Molecular Bioscience, The University of Sydney, Sydney, NSW 2006, Australia
B iThree Institute, University of Technology, Sydney, NSW, Australia
C Corresponding author. Tel: +61 2 9351 5383, Email: dee.carter@sydney.edu.au
Microbiology Australia 36(2) 93-95 https://doi.org/10.1071/MA15030
Published: 1 May 2015
Fungal organisms secrete a wide range of biomolecules, including degradative enzymes that are essential for nutrition, toxins, effectors and secondary compounds that modulate interactions with host animals and plants, and a variety of signaling and stress-related proteins1. As these are likely to be key determinants of virulence and may also be useful diagnostic and therapeutic targets, we investigated the secretome of different strains of the fungal pathogen Cryptococcus. Virulent strains secreted predominantly hydrolytic and proteolytic enzymes, while the least virulent strain secreted a range of additional non-degradative proteins including many that lacked secretion signals, some that appear to be ‘moonlighting’, and a number that are known to be allergenic. It appears that in Cryptococcus, the secretome may influence virulence both through the presence of harmful enzymes and through the absence of proteins that alert the host defence mechanisms.
Cryptococcus is an encapsulated yeast with two predominant pathogenic species: Cryptococcus neoformans and Cryptococcus gattii. These cause cryptococcosis in animals and humans, with disease ranging from asymptomatic to severe, fatal meningitis. There are a number of differences between C. gattii and C. neoformans including their preferred environmental niche, basidiospore morphology, drug susceptibility, epidemiology, the clinical manifestations of associated disease, and host susceptibility2. In addition, there are significant differences among stains within each species. In C. gattii, a hypervirulent sub-genotype designated VGIIa has caused a recent significant outbreak of cryptococcosis on Vancouver Island in British Columbia, Canada and in the Pacific Northwest of the United States. In contrast, a closely related sub-genotype designated VGIIb is globally distributed and hypovirulent3. These differences between Cryptococcus species and sub-genotypes provide an opportunity for understanding pathogenicity and disease progression by what are otherwise very genetically similar fungal organisms.
Our laboratory has been using ‘omics approaches to understand virulence in Cryptococcus, and used proteomic analysis to characterize the secretome produced by three Cryptococcus strains. Two strains were of high virulence (C. neoformans and C. gattii sub-genotype VGIIa) and the third of low virulence (C. gattii sub-genotype VGIIb). In our previous work on Cryptococcus proteomics, we found conditions optimized to simulate those encountered in the host induced the production of large amounts of shed capsular material, which interfered with the isolation and identification of proteins4. Therefore, we developed a novel method of protein capture using BioRad ProteoMiner™ beads, followed by mass spectroscopy. Sixty-seven cryptococcal proteins were identified and only one was common to all three strains. The secretomes of the high virulence C. neoformans and C. gattii VGIIa strains were similar and mostly consisted of a hydrolytic and proteolytic proteins. In contrast the lower virulence C. gattii VGIIb strain had a larger number of proteins with a greater diversity of functions (Figure 1). A significant proportion of these proteins are known to have roles in metabolism, signaling/transport, glycolysis and redox processes, and are considered to be canonical intracellular proteins. Published studies have reported very similar proteins in the extracellular milieu of various cell types from other organisms, and there is growing evidence that these may have ‘moonlighting’ functions, where they participate in completely different processes in alternative environments5. An additional subset of proteins found only in the C. gattii VGIIb secretome were orthologous to proteins known to elicit an immune response in the host, including the glycolytic proteins enolase and glyceraldehyde-3-phosphate dehydrogenase6,7. Most of these unusual secreted proteins lack secretion signals and are likely to be exported via alternative secretion pathways such as inside microvesicles, which have previously been isolated from Cryptococcus8; indeed the regulatory 14-3-3 protein, which is considered a biomarker of microvesicles9, was present exclusively in the VGIIb secretome.
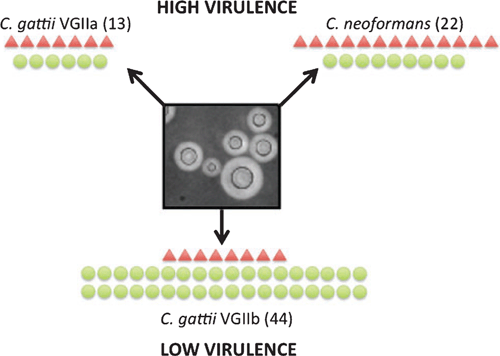
|
Mammals have a high level of innate immunity to most fungi, and the ability to infect immunocompetent hosts remains a rare trait. Cryptococcus is an environmental fungus, and as it cannot be spread from host to host, mammalian infection is likely to be accidental10. The questions of what determines virulence, and what processes underlie the evolution of strains that cause significant outbreaks in a dead-end host, are therefore intriguing. Comparative genomic studies have identified genes that are particular to the high virulence strains but their role in virulence is yet to be verified11,12. As secreted biomolecules are mediators of contact between the host and the pathogen, differences in these are likely to influence whether a pathogen will be rapidly recognized and eliminated, or will be able to bypass the host response and use host resources to establish an active infection. The results of our secretome analysis suggest that virulence in Cryptococcus may in part be determined by restricted secretion of proteins likely to elicit an immune response, and that in the absence of these the production and secretion of degradative enzymes enables host invasion.
References
[1] Girard, V. et al. (2013) Secretomes: the fungal strike force. Proteomics 13, 597–608.| Secretomes: the fungal strike force.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtlagsbo%3D&md5=48af3b43644bbc24c95823263c372f76CAS | 23349114PubMed |
[2] Springer, D.J. and Chaturvedi, V. (2010) Projecting global occurrence of Cryptococcus gattii. Emerg. Infect. Dis. 16, 14–20.
| Projecting global occurrence of Cryptococcus gattii.Crossref | GoogleScholarGoogle Scholar | 20031037PubMed |
[3] Byrnes, E.J. et al. (2010) Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6, e1000850.
| Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States.Crossref | GoogleScholarGoogle Scholar | 20421942PubMed |
[4] Chong, H.S. et al. (2012) Time-course proteome analysis reveals the dynamic response of Cryptococcus gattii cells to fluconazole. PLoS ONE 7, e42835.
| Time-course proteome analysis reveals the dynamic response of Cryptococcus gattii cells to fluconazole.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFOltrbK&md5=a31809a706bff0052428958892d10207CAS | 22880118PubMed |
[5] Huberts, D.H. and van der Klei, I.J. (2010) Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta 1803, 520–525.
| Moonlighting proteins: an intriguing mode of multitasking.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXjt1Gks7w%3D&md5=2c2d7542588777965990dd2b1b8ef956CAS | 20144902PubMed |
[6] Eroles, P. et al. (1997) The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins. Microbiology 143, 313–320.
| The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXht1entbc%3D&md5=257be4e6bc200d88f6a870c864198399CAS | 9043108PubMed |
[7] Gil-Navarro, I. et al. (1997) The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179, 4992–4999.
| 1:CAS:528:DyaK2sXlsVSmt7g%3D&md5=c9f16799a303b2b12fb0e2c3ae2c10c0CAS | 9260938PubMed |
[8] Rodrigues, M.L. et al. (2008) Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 7, 58–67.
| Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXkvVGjtLw%3D&md5=7c0c5f1e7ccfc8dbd406ad7126a6c1f2CAS | 18039940PubMed |
[9] Huang, S.-H. et al. (2012) Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS ONE 7, e48570.
| Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhslehsrnF&md5=633fc6b9777857c6813ff2d825cf4000CAS | 23144903PubMed |
[10] Casadevall, A. et al. (2003) ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi - the Cryptococcus neoformans paradigm. Curr. Opin. Microbiol. 6, 332–337.
| ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi - the Cryptococcus neoformans paradigm.Crossref | GoogleScholarGoogle Scholar | 12941400PubMed |
[11] Engelthaler, D.M. et al. (2014) Cryptococcus gattii in North American Pacific northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio 5, e01464-14.
| Cryptococcus gattii in North American Pacific northwest: whole-population genome analysis provides insights into species evolution and dispersal.Crossref | GoogleScholarGoogle Scholar | 25028429PubMed |
[12] Billmyre, R.B. et al. (2014) Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio 5, e01494-14.
| Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution.Crossref | GoogleScholarGoogle Scholar | 25073643PubMed |
Biographies
Leona Campbell has spent the past 13 years being fascinated by the fungal pathogen Cryptococcus, both as a PhD student and Postdoctoral researcher, primarily in Dee Carter’s lab. Her major area of interest is investigating host-pathogen interactions using ‘omics’ approaches. Leona also enjoys her teaching role overseeing the running of intermediate undergraduate Microbiology practical courses at the University of Sydney. She loves having the opportunity to inspire, and be inspired by, our next generation of microbiologists.
Dr Matt Padula is a Lecturer in the School of Biological Sciences and Professional Officer in the Proteomics Core Facility at the University of Technology Sydney. His research lies in the proteomic analysis of a range of organisms such as bacteria, yeast, mammalian tissue and cells, plant tissue, parasites, paralysis ticks, coral, snake venom and the pathogenic fungus Cryptococcus.
Liz Harry is a Professor of Biology and Deputy Director of the ithree institute (infection, immunology and innovation) at the University of Technology, Sydney (UTS). Liz obtained her PhD at the University of Sydney, was an NIH Fellow at Harvard, an Australian Research Council (ARC) Postdoctoral Fellow and an ARC QEII Fellow in the School of Molecular Biosciences at the University of Sydney. She has won an Australian Eureka Prize for Scientific research, and an ASM Frank Fenner Award. Her research focuses on bacterial cell division and antibacterials.
Dee Carter is an Associate Professor and head of the Discipline of Microbiology in the School of Molecular Bioscience, The University of Sydney, where she teaches mycology, medical microbiology and molecular biology. Her current research interests focus on using ‘omics approaches to understand fungal pathogenesis and to develop novel antifungal agents. She loves fungi because they are so adaptable and clever, making them excellent pets but also devastating enemies. She is particularly fond of Saccharomyces because it fits into the former category, Cryptococcus because it fits into the latter, and Aspergillus because it manages to straddle both.


