Integrative taxonomy of the new millipede genus Coxobolellus, gen. nov. (Diplopoda : Spirobolida : Pseudospirobolellidae), with descriptions of ten new species
Piyatida Pimvichai A F , Henrik Enghoff B , Somsak Panha C and Thierry Backeljau D E
A F , Henrik Enghoff B , Somsak Panha C and Thierry Backeljau D E
A Department of Biology, Faculty of Science, Mahasarakham University, Mahasarakham 44150, Thailand.
B Natural History Museum of Denmark, University of Copenhagen, Universitetsparken 15, DK-2100 Copenhagen Ø, Denmark.
C Animal Systematics Research Unit, Department of Biology, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand.
D Royal Belgian Institute of Natural Sciences, Vautierstraat 29, B-1000 Brussels, Belgium.
E Evolutionary Ecology Group, University of Antwerp, Universiteitsplein 1, B-2610 Antwerp, Belgium.
F Corresponding author. Email: piyatida.p@msu.ac.th
Invertebrate Systematics 34(6) 591-617 https://doi.org/10.1071/IS20031
Submitted: 16 April 2020 Accepted: 6 May 2020 Published: 14 August 2020
Journal Compilation © CSIRO 2020 Open Access CC BY-NC-ND
Abstract
Pseudospirobolellidae is a poorly known family of spirobolidan millipedes with only two genera and five described species. Yet, the descriptive taxonomy and molecular systematics of this group have been largely neglected. Therefore, the present work presents an integrative taxonomic study of new pseudospirobolellid taxa in Thailand. To this end, two mitochondrial gene fragments (COI and 16S rRNA) combined with morphological characters were used to define the genus Coxobolellus, gen. nov. with 10 new species, viz. C. albiceps, sp. nov., C. compactogonus, sp. nov., C. fuscus, sp. nov., C. nodosus, sp. nov., C. serratus, sp. nov., C. simplex, sp. nov., C. tenebris, sp. nov., C. tigris, sp. nov., C. transversalis, sp. nov. and C. valvatus, sp. nov. The interspecific COI sequence divergences among the new species ranged from 6 to 15%. The intergeneric COI sequence divergence between species of Coxobolellus, gen. nov., Benoitolus birgitae and Pseudospirobolellus sp. ranged from 20 to 23%. Three major morphological differences separate Coxobolellus, gen. nov. from Benoitolus and Pseudospirobolellus, namely (1) the protruding process on the 3rd (and 4th) coxae on male legs, (2) the posterior gonopod telopodite divided into two parts, and (3) a conspicuous opening pore at the mesal margin at the end of the coxal part of the posterior gonopod. Thus, the new genus is well supported by both mtDNA and morphological evidence, while the delimitation of the 10 new species is supported by the congruence between mtDNA and morphological data. Yet, with respect to the relationships of Benoitolus birgitae, morphological data suggest a similarity with Coxobolellus, gen. nov. and Pseudospirobolellus, whereas mtDNA data place this species in the Pachybolidae. Further phylogenetic analyses are needed to explore this apparent incongruence and test the monophyly of Pseudospirobolellidae.
Additional keywords: mitochondrial DNA, new genus, phylogeny, species delineation.
Introduction
The millipede order Spirobolida is one of the most diverse groups of Diplopoda, with ~1200 described species worldwide (Enghoff et al. 2015). Yet, this figure is steadily increasing, particularly with the recent discovery of new species of the family Pachybolidae in Thailand and Laos (e.g. Pimvichai et al. 2018). The present paper aims to describe another series of new spirobolidan taxa from south-east Asia. It focuses on the family Pseudospirobolellidae Brölemann, 1913, a species-poor and largely neglected taxon that currently comprises only two genera, namely Pseudospirobolellus Carl, 1912 (two species) and Benoitolus Mauriès, 1980 (three species) (Jeekel 2001; Enghoff et al. 2015). Several of these species were described from South-east Asia, a region that is a hotspot for diplopod diversity, such that each time taxonomists start working on a particular family, the numbers of new species rise sky high (e.g. 35 species of the Harpagophoridae (e.g. Pimvichai et al. 2016) and 68 species of the Paradoxosomatidae (e.g. Likhitrakarn et al. 2011; Srisonchai et al. 2018a, 2018b)). This turns out to apply also to the Pseudospirobolellidae, as will be illustrated with this paper in which 10 new species and one new genus of this family are described from Thailand by combining morphological and DNA sequence data.
Material and methods
Live specimens were hand collected in the field (for locality data see Table 1). Some were preserved in 70% ethanol for morphological studies, whereas the rest were placed in a freezer at –20°C for subsequent DNA studies. Specimens from the following collections were also examined:
-
CUMZ, Museum of Zoology, Chulalongkorn University, Bangkok, Thailand.
-
NHMW, Naturhistorisches Museum, Vienna, Austria.
-
NHMD, Natural History Museum of Denmark, University of Copenhagen, Denmark.
This research was conducted under the approval of the Animal Care and Use regulations (numbers U1-07304-2560 and IACUC-MSU-037/2019) of the Thai government.
Morphology
Gonopods were photographed with a digital camera manipulated via the program Helicon Remote (ver. 3.1.1.w). The Zerene Stacker Pro software was used for image-stacking. Samples for scanning electron microscopy (SEM) were air-dried directly from alcohol and sputter-coated for 250 s with gold. Scanning electron micrographs were taken with an environmental scanning electron microscope (ESEM)-FEI Quanta 200. Drawings were made using a stereomicroscope and photographs. The identification and classification of the specimens followed Carl (1912), Attems (1936, 1953), Mauriès (1980) and Hoffman (1981). Voucher specimens were deposited in the collection of CUMZ.
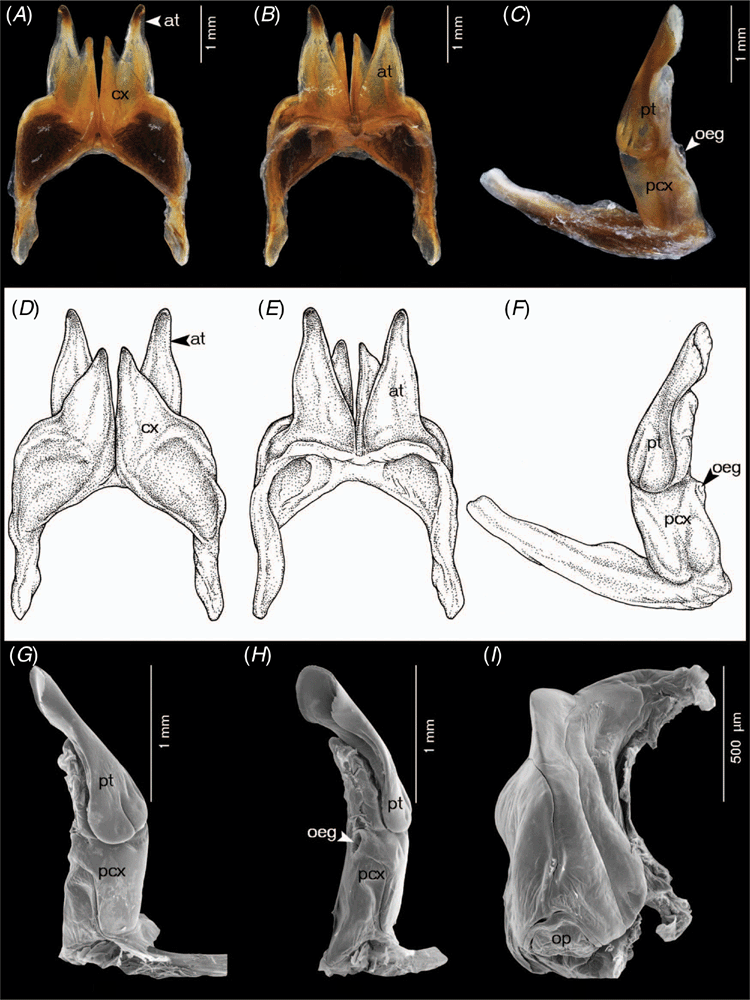
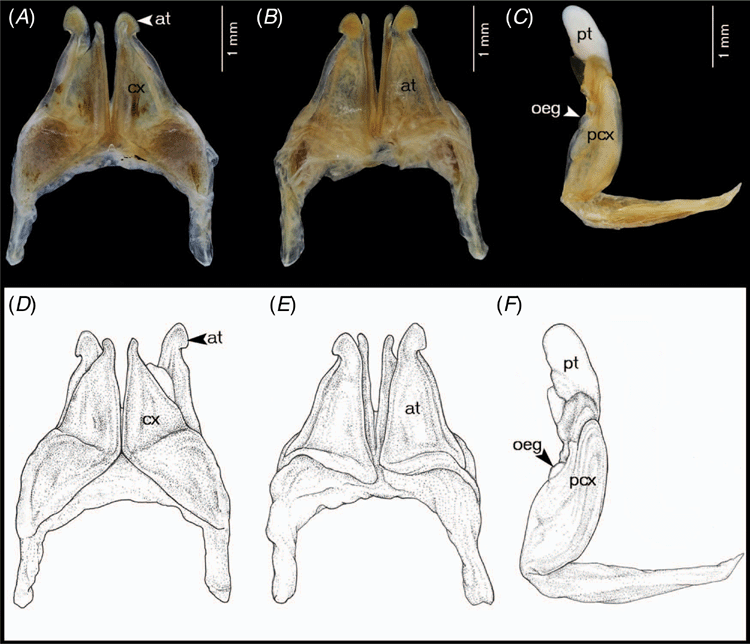
Terminology and abbreviations for the structure of gonopods and vulvae in Coxobolellus, gen. nov.
-
at, telopodite of the anterior gonopod, distinctly seen in posterior view
-
cx, coxal part of anterior gonopod
-
oeg, opening of efferent groove
-
pcx, coxal part of posterior gonopod
-
pt, telopodital part of the posterior gonopod
-
op, operculum of female vulvae
DNA extraction, amplification and sequencing
Total genomic DNA was extracted from dissected legs using the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions. PCR amplifications were done in 11-µL reaction volumes containing 5.50 µL of Multiplex PCR Master Mix (Qiagen), 1 µL of each primer, 2.5 µL of sterilised distilled water and 1 µL of DNA extract. Two mitochondrial DNA gene fragments (COI and 16S rRNA) were amplified with the primers LCO-1490 and HCO-2198 (Folmer et al. 1994) for COI, and 16Sar and 16Sbr (Kessing et al. 2004) for 16S rRNA. The PCR conditions for the amplification of COI and 16S rRNA were: initial denaturation at 95°C for 15 min, followed by 36 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 1 min, and then a final extension at 72°C for 10 min. PCR amplification success was evaluated by comparing the amplicon sizes in 5 µL of PCR product through electrophoresis on 1% (w/v) agarose–TBE gel, stained with SYBR Safe and visualised by UV trans-illumination. The remaining portion of each PCR reaction (5 µL) was purified with the ExoSAP protocol (with 37°C for 15 min and 85°C for 15 min). DNA sequencing was performed with the BigDye Terminator (ver. 1.1) cycle Sequencing kit (Applied Biosystems, Lennik, Belgium), using the PCR primers. Sequencing was done with an ABI 3130xl capillary DNA sequencer (Applied Biosystems).
The COI data included 48 specimens, representing 19 genera and 13 nominal species of ingroup taxa (Table 1). The 16S rRNA data included 42 specimens, i.e. the same specimens as for COI, plus Litostrophus scaber (Verhoeff, 1938) and Trachelomegalus cf. hoplurus, but minus Benoitolus birgitae (Hoffman, 1981), Coxobolellus albiceps, sp. nov. (Stpl), C. simplex, sp. nov. (TNP) and C. valvatus, sp. nov. (TCD) in the ingroup and Paraspirobolus lucifugus (Gervais, 1836) and Narceus annularis Rafinesque, 1820 in the outgroup. Thus, the combined COI + 16S rRNA data included 40 specimens. Species of the order Spirostreptida, namely Anurostreptus barthelemyae Demange, 1961 (Harpagophoridae), Chonecambala crassicauda Mauriès & Enghoff, 1990 (Pericambalidae) and Thyropygus allevatus (Karsch, 1881) (Harpagophoridae) were used as outgroup for tree reconstruction. All nucleotide sequences have been deposited in GenBank under accession numbers MT328211–MT328227 and MT328992–MT329012 for the partial 16S rRNA and COI fragment sequences respectively. Sample data and voucher codes are shown in Table 1.
Alignment and phylogenetic analysis
CodonCode Aligner (ver. 4.0.4, CodonCode Corporation) was used to assemble the forward and reverse sequences and to check for errors and ambiguities. The COI and 16S rRNA sequences were checked with the Basic Local Alignment Search Tool (BLAST) provided by NCBI and compared with reference sequences in GenBank. They were aligned using MUSCLE (ver. 3.6, see http://www.drive5.com/muscle; Edgar 2004). The alignments consisted of 660 bp for COI and 458 bp for 16S rRNA (gaps were excluded by complete-deletion). The sequences were checked for ambiguous nucleotide sites, saturation and phylogenetic signal using DAMBE (ver. 5.2.65. see http://www.dambe.bio.uottawa.ca/DAMBE/dambe.aspx; Xia 2018). MEGA (ver. X, see http://www.megasoftware.net; Tamura et al. 2013; Kumar et al. 2018) was used to (1) translate COI protein-coding sequences into amino acids, (2) check for stop codons, (3) calculate uncorrected pairwise p-distances among sequences, and (4) evaluate transition/transversion ratios.
Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian inference (BI). The shape parameter of the gamma distribution, based on 16 rate categories, was estimated using maximum-likelihood analysis. ML trees were inferred with RAxML (ver. 8.2.12, see http://www.phylo.org/index.php/tools/raxmlhpc2_tgb.html; Stamatakis 2014) through the CIPRES Science Gateway (Miller et al. 2010) using a GTR+G substitution model and 1000 bootstrap replicates to assess branch support. The COI sequence alignment was partitioned by 1st, 2nd and 3rd codon position and the concatenated sequence alignment was partitioned by gene fragment and by 1st, 2nd and 3rd codon position for COI.
BI trees were constructed with MrBayes (ver. 3.1.2, see http://www.phylo.org/index.php/tools/mrbayes_xsede.html; Huelsenbeck and Ronquist 2001) for COI and 16S rRNA separately, as well as for the combined data. Substitution models were inferred separately for each gene fragment using PartitionFinder 2 on XSEDE (ver. 2.1.1, see http://www.phylo.org/index.php/tools/partitionfinder2_xsede.html; Lanfear et al. 2017) through the CIPRES Science Gateway (Miller et al. 2010). BI trees were run for 2 million generations (heating parameters were 0.05 for COI and 0.07 for 16S rRNA and combined datasets), sampling every 1000 generations. Convergences were confirmed by verifying that the standard deviations of split frequencies were below 0.01. Then the first 1000 trees were discarded as burn-in, so that the final consensus tree was built from the last 3002 trees. The optimal parameters were assessed by calculating the Bayes factor using Tracer (ver. 1.6.0, A. Rambaut, M. A. Suchard, W. Xie, and A. J. Drummond, see http://beast.bio.ed.ac.uk/Tracer, accessed 11 December 2019). Support for nodes was assessed by posterior probabilities.
We consider clades with bootstrap values (BV) of ≥70% to be well supported (Hillis and Bull 1993) and <70% as poorly supported. For BI analyses, we considered clades with posterior probabilities (PP) of ≥0.90 (e.g. Shiels et al. 2014) to be well supported and <0.90 as poorly supported.
Species delimitation
Putative species were explored by applying the General Mixed Yule Coalescent (GMYC) (Fujisawa and Barraclough 2013) and the Automatic Barcode Gap Discovery (ABGD) (Puillandre et al. 2012) methods to the COI sequence data, i.e. the standard DNA barcoding fragment (Hebert et al. 2003). GMYC requires an ultrametric gene tree (fully dichotomous tree) input to derive a BI tree using BEAST (ver. 1.8.2, see http://www.tree.bio.ed.ac.uk/software/beast/; Drummond et al. 2012). An XML file was made with the BEAUti (ver. 1.8.2, see http://www.beast.community/) interface under a lognormal relaxed (uncorrelated) molecular clock with the GTR substitution model. Putative species were identified using the single and multiple GMYC thresholds via the web interface at https://species.h-its.org/gmyc/.
With ABGD, barcode gaps can be observed whenever the sequence divergence among conspecific specimens is smaller than the sequence divergence among specimens from different species (http://wwwabi.snv.jussieu.fr/public/abgd/). COI alignments were uploaded to the ABGD server at http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html and were run with the default settings, except for the relative gap width, which was set at X = 0.98 (0.98 was the highest value that could be applied because the default value for relative gap width (X = 1.5) value did not produce a result for the dataset). COI sequence divergence was estimated with K2P distances.
Results
The nucleotide frequencies in the aligned COI gene fragment (660 bp) were: A, 0.284; C, 0.208; G, 0.159; T, 0.349 – 36.7% GC content. The uncorrected p-distance between the sequences ranged from 0.00 to 0.26 (Table 2).

|
The nucleotide frequencies in the aligned 16S rRNA gene fragment (458 bp) were: A, 0.325; C, 0.108; G, 0.212; T, 0.355 – 31.9% GC content. The uncorrected p-distance between the sequences ranged from 0.00 to 0.29 (Table 3).

|
The nucleotide frequencies in the COI + 16S rRNA dataset (1118 bp) were: A, 0.302; C, 0.166; G, 0.180; T, 0.352 – 34.6% GC content. The uncorrected p-distance between the sequences ranged from 0.00 to 0.27.
Phylogeny
The ML and BI trees for the separate and combined datasets (COI, 16S rRNA and COI + 16S rRNA) were largely congruent (by visual inspection of the branching pattern). The single 16S rRNA and the combined (COI + 16S rRNA) trees are presented in the Supplementary material (Fig. S1, S2 respectively), whereas the COI tree is used for further discussion (Fig. 1). PartitionFinder indicated that the best substitution models for BI analysis were GTR+I+G for 16S rRNA and the 1st and 2nd codon positions of COI, and GTR+G for the 3rd codon position.
Although the COI tree includes representatives of six Spirobolida families (Pachybolidae, Pseudospirobolellidae, Rhinocricidae, Spirobolidae, Spirobolellidae and Trigoniulidae), it provides no meaningful support for the relationships and monophyly of the family Pachybolidae. The three trigoniulid genera Apeuthes Hoffman & Keeton, 1960, Leptogoniulus Silvestri, 1897 and Trigoniulus Pocock, 1894 form a well supported clade (BV = 97; PP = 1.00).
Benoitolus birgitae groups with Litostrophus (Pachybolidae) and thus is well separated from the genus Pseudospirobolellus. However, this is based on COI only and with a fairly long branch, the position of which in the tree is ambiguously supported. COI sequence divergence between B. birgitae and (1) Coxobolellus, gen. nov. is 20–23% (mean: 22%), (2) Pseudospirobolellus is 23%, and (3) Pachybolidae genera is 11–24% (mean: 21%). The lowest COI sequence divergence is with Litostrophus (11–18%; mean: 15%).
The sister-group relation between Coxobolellus, gen. nov. and Pseudospirobolellus is well supported by both ML and BI in all trees, just as is the sister-group relation of the two Pseudospirobolellus species.
Clade 1 is well supported and comprises 18 sequences of 10 Coxobolellus, gen. nov. species that are grouped into four well supported clades (1A–D) (except for Clade 1B in the 16S rRNA tree, Fig. S1).
Clade 1A comprises C. nodosus, sp. nov. and C. valvatus, sp. nov., two species that are distributed in western and northern Thailand respectively. Their anterior gonopods are similar, but they differ in their posterior gonopod telopodite, and their COI sequence divergence is 6–7%.
Clade 1B comprises two sympatric species: C. albiceps, sp. nov. and C. transversalis, sp. nov. They differ in the tip of the anterior gonopod and the posterior gonopod telopodite, and show 8% COI sequence divergence.
Clade 1C comprises C. compactogonus, sp. nov. and C. tenebris, sp. nov., which are distributed in north-eastern and central to western Thailand respectively. They differ in both the anterior gonopod and posterior gonopod telopodite, and their COI sequence divergence is 9–10%.
Clade 1D comprises C. serratus, sp. nov. and C. tigris, sp. nov., which have similar anterior gonopods, but differ in their posterior gonopod telopodite. Both species are distributed in southern Thailand, and their COI sequence divergence is 12%.
Species delimitation based on COI sequences
GMYC
With both the single and multiple thresholds, the maximum-likelihood values of the null model (=all sequences belong to a single species) were lower than the maximum-likelihood value of the GMYC model. The single threshold yielded 4 clusters and 11 entities, whereas the multiple thresholds yielded 4 clusters and 7 entities (Table 4).

|
ABGD
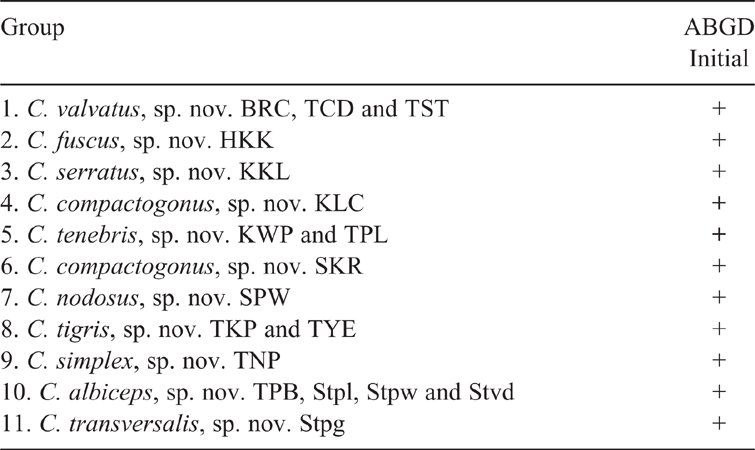
This method was run with prior intraspecific K2P divergences ranging from Pmin = 0.001 to Pmax = 0.1. The initial partition of sequences into 11 operational taxonomic units (OTUs) based on a statistically inferred barcode gap (Kekkonen et al. 2015) was stable over the range of prior values of intraspecific K2P sequence divergence (0.001–0.100), such that the same 11 OTUs were consistently found (Table 5).
|
The K2P intraspecific COI sequence divergences of Coxobolellus species ranged from 0.00 to 0.05. Therefore, in this study COI K2P sequence divergences above 0.05, accompanied by distinct gonopodal characters, are regarded as indicative of species-level differentiation. As such, congeneric interspecific COI K2P sequence divergences ranged from 0.06 to 0.15, with an average of 11% (Table 2), whereas intergeneric COI K2P sequence divergences between the genera Coxobolellus, gen. nov. and Pseudospirobolellus ranged from 0.20 to 0.23.
Taxonomic corollary
Coxobolellus, gen. nov. includes 10 well supported new species which share three synapomorphies:
-
posterior gonopod telopodite clearly divided into two parts: coxal part (pcx) and telopodital part (pt);
-
coxae of the 3rd (and 4th) leg with extremely large, protruding process;
-
opening pore (oeg) at mesal margin at the end of coxal part (pcx).
These three characters are autapomorphic for Coxobolellus, gen. nov. In the next section we provide formal descriptions of these new taxa.
Taxonomy
Class DIPLOPODA de Blainville in Gervais, 1844
Order SPIROBOLIDA Bollman, 1893
Suborder SPIROBOLIDEA Bollman, 1893
Family Pseudospirobolellidae Brölemann, 1913
A family of the order Spirobolida characterised, inter alia, by the strongly to almost completely reduced anterior gonopod sternum (Pitz and Sierwald 2010).
Fifteen species are included:
-
Genus Pseudospirobolellus Carl, 1912: P. avernus (Butler, 1876); P. sigmoides Attems, 1953.
-
Genus Benoitolus Mauriès, 1980 (=Solaenobolellus Hoffman, 1981): B. flavicollis Mauriès, 1980; B. birgitae (Hoffman, 1981); B. siamensis (Attems, 1936) (=Cyclothyrophorus siamensis Attems, 1936) (see Jeekel 2001, p. 47).
-
Genus Coxobolellus, gen. nov.: C. albiceps, sp. nov.; C. compactogonus, sp. nov.; C. fuscus, sp. nov.; C. nodosus, sp. nov.; C. serratus, sp. nov.; C. simplex, sp. nov.; C. tenebris, sp. nov.; C. tigris, sp. nov.; C. transversalis, sp. nov.; C. valvatus, sp. nov.
Pitz and Sierwald (2010) included two undescribed species of Pseudospirobolellidae from China in their analysis, which suggests that there remains taxonomic diversity to be discovered in this family outside Thailand.
Genus Coxobolellus, gen. nov.
(Fig. 2)
Type species: Coxobolellus tenebris, sp. nov.
Diagnosis
Differing from the other genera of Pseudospirobollelidae by having the coxae of the 3rd male leg-pair with extremely large, protruding processes (in C. albiceps, sp. nov. and C. transversalis, sp. nov., also the coxae of the 4th male leg-pair with extremely large, protruding processes). The 4th–5th leg-pairs with large, triangular coxae (Fig. 2A–C). Telson smooth; preanal ring with a short process protruding as far as or slightly beyond anal valves (Fig. 2D). Anal valves smooth, rounded. Subanal scale broadly triangular (Fig. 2E). Posterior gonopod telopodite divided into a coxal part (pcx) and a telopodital part (pt); with opening of efferent groove (oeg) at mesal margin at the end of coxal part (pcx).
General description
Adult males with 47–57 podous rings, length ~4–8 cm, diameter ~3.2–6.0 mm. Adult females with 47–57 podous rings, length ~5–9 cm, diameter ~4.1–6.4 mm. No apodous rings in front of telson, except where noted.
Head capsule smooth. Occipital furrow extending down between, but not beyond eyes; clypeal furrow reaching level of antennal sockets. Area below antennal sockets and eyes impressed, forming part of antennal furrow. Incisura lateralis open. 2+2 labral teeth, a row of labral setae, 2+2, 3+3 or 4+4 supralabral setae. Diameter of eyes approximately half of interocular space; 6–9 vertical rows of ommatidia, 4 or 5 horizontal rows, 24–32 ommatidia per eye. Antennae short, not reaching beyond collum when stretched back, accommodated in a shallow furrow composed of a horizontal segment in the head capsule and a vertical segment in the mandibular cardo and stipes. Antennomere lengths 2 > 1 > 6 > 3 > 4 = 5 > 7; antennomere 1 glabrous, 2 and 3 with some ventral setae, 4, 5 and 6 densely setose; 4 apical sensilla. Gnathochilarium: each stipes with 2 or 3 apical setae; each lamella lingualis with 2 setae, one behind the other. Basal part of mentum transversely wrinkled; basal part of stipes longitudinally wrinkled.
Collum smooth, with a marginal furrow along lateral part of anterior margin; lateral lobes narrowly rounded, extending as far ventrad as the ventral margin of body ring 2.
Body rings 2–5 ventrally concave, hence with distinct ventrolateral corners. Body rings very smooth, sides parallel in dorsal view. Tergo-pleural suture visible on pro- and mesozona. Prozona smooth; mesozona ventrally with fine oblique striae, dorsally punctate; metazona ventrally with fine longitudinal striae, otherwise smooth. Pleural parts of rings with fine oblique striae. Sterna transversely striate. Ozopores from ring 6, situated in mesozona, ~1 pore diameter in front of metazona. Sutures between pro-, meso- and metazona distinct; horizontal suture distinct on pro-, meso- and metazona.
Telson
Smooth, preanal ring with slightly concave dorsal profile, with a short process protruding as far as, or slightly beyond, anal valves. Anal valves smooth, rounded. Subanal scale broadly triangular.
Legs
Length of midbody legs 51–81% of body width in males, 48–70% of body width in females. Prefemur basally constricted and longer than other podomeres. First and second legs with 1 or 2 prefemoral, 1 femoral, 1 postfemoral, and 2 tibial setae, and 3 ventral and 1 dorsal apical setae on tarsi, numbers of setae reaching constancy from pair 3: each leg with 1 tibial seta; tarsi in males with 2 or 3 ventral apical and one dorsal apical seta; females with 1 coxal, 1 prefemoral, 1 femoral, 1 postfemoral, 1 tibial seta, tarsi with 1–3 ventral and one dorsal apical seta, the apical ventral seta larger than the more basal one.
Male sexual characters
Prefemur of all legs from third to last pair with large ventral soft pad (Fig. 2F, G) occupying entire ventral surface. Body ring 7 entirely fused ventrally, no trace of a suture. Anterior gonopods without a sternum, tip of anterior gonopods visible when the animal is stretched out (not when it is rolled up). Posterior gonopods in situ completely hidden within anterior ones. Posterior gonopod telopodite divided into a coxal part (pcx) and a telopodital part (pt); with opening of efferent groove (oeg) at mesal margin at the end of coxal part (pcx).
Female vulvae
Large, in situ projecting beyond lateral extensions of coxosternum of 2nd legs. Operculum small, rounded triangular, situated at laterobasal end of vulva. Shape of valves variable.
Distribution
Hitherto known only from Thailand
Ecology
Specimens of Coxobolellus gen. nov. are mostly found under leaf litter. Sometimes specimens are found inside rotten wood or climbing on trees.
Etymology
The name emphasises the importance of coxal characters in the diagnosis of the new genus.
Species descriptions
Coxobolellus albiceps, sp. nov.
Material examined
Holotype. Male, Thailand, Nan Province, Muang District, Tham Pha Tub, 18°51′17″N, 100°44′10″E, 9.vi.2019, P. Pimvichai and T. Backeljau leg. (CUMZ).
Paratypes. Thailand: 5 males, 6 females, same data as holotype. (CUMZ). 1 male, 2 females, Chiang-Rai Province, Pan District, Wat Tham Bampen Bun, 19°42′36″N, 99°45′13″E, 10.i.2008, P. Pimvichai leg. (CUMZ). 2 males, 2 females, Phrae Province, Rong Kwang District, Tham Pha Nang Khoi, 18°21′41″N, 100°21′03″E, 9.vi.2019, P. Pimvichai and T. Backeljau leg. (CUMZ). 4 males, 4 females, Phrae Province, Rong Kwang District, Huai Rong Waterfall, 18°26′31″N, 100°27′01″E, 31.viii.2014, P. Pimvichai leg. (CUMZ). 6 males, 5 females, 10.vi.2019, P. Pimvichai and T. Backeljau leg. (CUMZ). 5 males, 5 females, Phitsanulok Province, Noen Maprang, Tham Wang Daeng, 16°40′37″N, 100°41′38″E, 25.ix.2018, P. Pimvichai and S. Saratan leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the tip of the anterior gonopod coxa obliquely truncated, and by having the telopodital part (pt) of the posterior gonopod short compared with the coxal part (pcx).
Description
Adult males with 48–53 podous rings. Length ~3–6 cm, diameter ~3.8–4.7 mm (3.2–3.4 mm in small individuals). Adult females with 47–51 podous rings. Length ~3–6 cm, diameter ~4.3–4.8 mm (4.1 mm in small individuals).
Colour of living animal dark green. Specimens from Tham Wang Daeng and some specimens from Tham Pha Tub brown, metazona dark brown. Antennae, collum, legs and preanal process beige in all specimens. (Fig. 13A–C).
Anterior gonopods (Fig. 3A, B, D, E) with high coxae, apically obliquely truncated, posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite overreaching coxa, apically narrowly rounded with pigmented brown node, with constricted lateral margin at ⅔ of its height, forming a triangular process.
Posterior gonopods (Fig. 3C, F–I) very simple, with long, smooth coxal part (pcx); telopodital part (pt) curving mesad, flattened, concave forming a canopy, with strong transverse ridge near base, with a short spine protruding from surface near tip.
Female vulvae (Fig. 3J): valves prominent, of equal size.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT328994 (voucher code CUMZ-D00121).
Distribution
Nan, Phrae, Chiang-Rai and Phitsanulok provinces, Thailand (Fig. 14).
Note
This species is polymorphic for both colour pattern and size. Specimens are dark green, green or brown, but the antennae, collum, legs and preanal process are always beige. Brown specimens are smaller both in length and diameter than dark green or green specimens. The three morphs occur in Tham Pha Tub (Fig. 13A–C) and Huai Rong Waterfall, but in Tham Wang Daeng only the brown morph (length 3–4 cm, diameter 3.2–3.4 mm) was found.
Etymology
The specific epithet is a Latin noun in apposition, meaning ‘white/pale head’ and referring to the contrastingly pale head in living specimens (Fig. 13A–C).
Coxobolellus compactogonus, sp. nov.
Material examined
Holotype. Male, Thailand, Nakhon Ratchasima Province, Wang Nam Khiao District, Sakaerat Environmental Research Station, 14°30′36″N, 101°55′51″E, 24.iv.2009, P. Pimvichai leg. (CUMZ).
Paratypes. Thailand: 1 male, same data as holotype (CUMZ). 2 males, 2 females, Nakhon Ratchasima Province, Pak Chong District, Khao Look Chang, 14°31′34″N, 101°21′30″E, 24.vi.2008, P. Pimvichai leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the anterior gonopod telopodite (at) with a rounded tip projecting slightly over coxa (cx), and by having the telopodital part (pt) of the posterior gonopod short and compact, with three processes surrounding a deep concavity.
Description
Adult males with 47–55 podous rings, 1 specimen from Sakaerat and 1 specimen from Khao Look Chang with 2 apodous rings. Length ~5–6 cm, diameter ~4.1–4.6 mm. Adult females with 47–50 podous rings. Length ~7 cm, diameter ~4.8–5.2 mm.
Colour after 10 years in ethanol: head, antenna, collum, legs and telson light brown; body rings dark brown.
Anterior gonopods (Fig. 4A, B, D, E) with high, triangular coxae, gradually narrowing towards tip; at base of posterior surface with relatively high ridge laterally for accommodation of telopodite. Telopodite projecting slightly over anterior gonopod coxa (cx), simple, gradually narrow towards tip, apically rounded, curving mesad.
Posterior gonopods (Fig. 4C, F–I) simple, rounded, with very long, smooth coxal part (pcx); telopodital part (pt) short, with three processes, basal part and apical part ending in a rounded lobe, the lateral process with serrate margin. The three processes forming a deep concavity.
Female vulvae (Fig. 4J): valves prominent, of equal size.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT328998 (voucher code CUMZ-D00134).
Distribution
Known only from the type locality in Nakhon Ratchasima Province, Thailand (Fig. 14).
Etymology
The specific epithet is a noun in apposition referring to the particularly compact posterior gonopod.
Coxobolellus fuscus, sp. nov.
Material examined
Holotype. Male, Thailand, Kanchanaburi Province, Sangkhla Buri District, Kroeng Krawia waterfall, 14°58′51″N, 98°37′57″E, 8.vi.2010, P. Pimvichai leg. (CUMZ).
Paratypes. Thailand: 1 male, 2 females, same data as holotype (CUMZ). 3 males, 1 female, Kanchanaburi Province, Thong Pha Phum District, Wat Tha Khanun, 14°44′30″N, 98°38′14″E, 24.vii.2016, P. Pimvichai and T. Backeljau leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the tip of the anterior gonopod coxa ending in an abruptly narrowed, long process, and by having the tip of the telopodital part (pt) of the posterior gonopod ending in a coarsely serrate lamella with a sharp point.
Description
Adult males with 50–57 podous rings, 1 specimen from Wat Tha Khanun with 3 apodous rings. Length ~6–7 cm, diameter ~3.8–4.3 mm. Adult females with 51–57 podous rings. Length ~7 cm, diameter ~4.5–4.6 mm.
Colour of living animal: antennae, legs and metazona reddish brown, collum and prozona dark brown, preanal process yellowish brown (Fig. 13D).
Anterior gonopods (Fig. 5A, B, D, E) with very high coxae, gradually narrowing towards tip, apically slender, narrow, mesal margin straight; at base of posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite far overreaching coxa, apically abruptly narrow, curving mesad, with constricted lateral margin at ~⅔ of its height, forming a triangular process.
Posterior gonopods (Fig. 5C, F–I) simple, rounded, with very long, smooth coxal part (pcx); telopodital part (pt) short, curving mesad, apically ending in a coarsely serrate lamella with a sharp point.
Female vulvae (Fig. 5J): valves prominent, of equal size.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT328999 (voucher code CUMZ-D00133).
Distribution
Kanchanaburi Province, Thailand (Fig. 14).
Etymology
The specific name is a Latin adjective, meaning ‘brown’ and referring to the general body colour of living specimens (Fig. 13D).
Coxobolellus nodosus, sp. nov.
Material examined
Holotype. Male, Thailand, Tak Province, Mae Sot District, Chao Por Phawo Shrine, 16°40′17″N, 98°41′08″E, 17.vii.2008, P. Pimvichai leg. (CUMZ).
Paratypes. Thailand: 5 males, 3 females, 2 juveniles, same data as holotype (CUMZ).
Diagnosis
Differing from all other species in the genus by having the telopodital part (pt) of the posterior gonopod ending in a long, sharp spine, and by having a flattened lamella protruding from the mesal surface near the tip. Further differing from all other species, except C. valvatus, sp. nov., by having the tip of the anterior gonopod coxa concave.
Description
Adult males with 50–52 podous rings. Length ~7–8 cm, diameter ~5.3–5.5 mm. Adult females with 49–51 podous rings. Length ~8–9 cm, diameter ~5.6–6.0 mm.
Colour after 11 years in ethanol: head, antenna, legs and telson reddish brown; body rings dark brown.
Anterior gonopods (Fig. 6A, B, D, E) with high coxae, apically narrowly concave, mesal margins straight, diverging, delimiting a V-shaped space between both coxae, posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite slightly overreaching coxa, apically narrow, forming a triangular process with a pigmented brown node, with constricted lateral margin at ~⅔ of its height, forming a triangular process.
Posterior gonopods (Fig. 6C, F–H) simple, with long, smooth coxal part (pcx); telopodital part (pt) as long as coxal part, curving mesad, forming a deep concavity, mesal margin forming a broadly expanded sheet, with serrate mesal margin; apically ending in a long, sharp spine, with a flattened lamella at base of apical sharp spine, protruding from mesal surface.
Female vulvae (Fig. 6I): valves prominent, of equal size; free margins meeting in sigmoid suture.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329000 (voucher code CUMZ-D00126).
Distribution
Known only from the type locality in Tak Province, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin adjective referring to the pigmented node at the tip of the anterior gonopod telopodite of this species.
Coxobolellus serratus, sp. nov.
Material examined
Holotype. Male, Thailand, Prachuap Khiri Khan Province, Pran Buri District, Khao Kalok, 12°20′04″N, 99°59′50″E, 13.x.2008, C. Sutcharit leg. (CUMZ).
Paratypes. Thailand: 1 male, same data as holotype (CUMZ).
Diagnosis
Differing from all other species in the genus by having the anterior gonopod telopodite (at) far overreaching coxa (cx) and curving laterad, and by having a lateral serrate margin on the telopodital part (pt) of the posterior gonopod.
Description
Adult males 52 podous rings, 1 specimen with 2 apodous rings. Length ~6–7 cm, diameter ~4.4–4.9 mm.
Colour after 11 years in ethanol: brown; antenna, legs and metazona dark brown.
Anterior gonopods (Fig. 7A, B, D, E) with high, triangular coxae, lateral margin concave at 1/2 of its height, mesal margin straight; at base of posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite far overreaching coxa, apically abruptly narrow, curving laterad.
Posterior gonopods (Fig. 7C, F–H) very simple, with long, smooth coxal part (pcx); telopodital part (pt) as long as pcx, curving mesad, laterally with serrate margin.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329001 (voucher code CUMZ-D00132).
Distribution
Known only from the type locality in Prachuap Khiri Khan Province, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin adjective referring to the prominent serration of the telopodital part of the posterior gonopod.
Coxobolellus simplex, sp. nov.
Material examined
Holotype. Male, Thailand, Lampang Province, Mae Prik District, Tham Pha Pha Ngam, 17°28′49″N, 99°10′5″E, 7.viii.2008, C. Sutcharit leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the telopodital part (pt) of the posterior gonopod short, with a flattened lamella at base, apically ending in a rounded lobe. Further differing from the other species, except C. tenebris, sp. nov., by having the tip of the telopodital part (pt) curving mesad, rounded, smooth.
Description
Adult male with 52 podous rings. Length ~6 cm, diameter ~4.6 mm.
Colour after 11 years in ethanol: collum and telson orange-brown; prozona grey; body ring and legs light brown.
Anterior gonopods (Fig. 8A, B, D, E) with high, triangular coxae, apically abruptly narrower, pointing mesad, lateral margin concave at ½ of its height, mesal margin straight; at base of posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite far overreaching coxa, apically rounded, with constricted lateral margin near tip, forming a triangular process.
Posterior gonopods (Fig. 8C, F) simple, rounded, with very long, smooth coxal part (pcx); telopodital part (pt) short, with a flattened lamella at base, apically ending in a rounded lobe.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329002 (voucher code CUMZ-D00136).
Distribution
Known only from the type locality in Lampang Province, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin adjective referring to the particularly simple posterior gonopod.
|
Coxobolellus tenebris, sp. nov.
Material examined
Holotype. Male, Thailand, Uthai Thani Province, Ban Rai District, Wat Khao Wong Phrohm-majan, 15°18′09″N, 99°45′16″E, 8.vii.2009, P. Pimvichai leg. (CUMZ).
Paratypes. Thailand: 3 males, 2 females, same data as holotype (CUMZ). 1 male, Kanchanaburi Province, Sai Yok District, Wat Tham Phrom Lok Khao Yai, 14°12′16″N, 99°07′58″E, 9.vii.2009, P. Pimvichai leg. (CUMZ). 2 males, 1 female, Suphan Buri Province, Dan Chang District, Wat Weluwan Khiriwong, 14°46′05″N, 99°26′33″E, 8.vii.2009, P. Pimvichai leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the tip of the anterior gonopod coxa very strongly narrowed and pointed, and by having a slender triangular lamella curling around the basal part of the telopodital part (pt) of the posterior gonopod. Further differing from all other species except for C. simplex, sp. nov. from Tham Pha Pha Ngam in having the tip of the telopodital part (pt) curving mesad, rounded, and smooth.
Description
Adult males with 53–55 podous rings, 1 specimen with 2 apodous rings. Length ~7–8 cm, diameter ~5.0–6.0 mm. Adult females with 48–53 podous rings. Length ~8–9 cm, diameter ~5.6–6.4 mm.
Colour of living animal: dark greenish grey. Edge of metazona and preanal process light brown (Fig. 13E).
Anterior gonopods (Fig. 3A, B, D, E) with high, triangular coxae, apically strongly narrowed, pointed. Telopodite far overreaching coxa, apically narrowly rounded, with a tiny triangular laterad denticle near tip.
Posterior gonopods (Fig. 3C, F–I) very simple, with very long, smooth coxal part (pcx); telopodital part (pt) curving mesad, rounded, smooth, with a slender triangular lamella curling around basal part.
Female vulvae (Fig. 3J): valves prominent, of equal size.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329004 (voucher code CUMZ-D00120).
Distribution
Uthai Thani, Kanchanaburi and Suphan Buri provinces, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin adjective, meaning ‘dark’ and referring to the general body colour of living specimens.
Coxobolellus tigris, sp. nov.
Material examined
Holotype. Male, Thailand, Chumphon Province, Pathio District, Tham Yai I, 10°44′45″N, 99°23′45″E, 22.v.2010, P. Pimvichai leg. (CUMZ).
Paratypes. Thailand: 2 males, 4 females, same data as holotype (CUMZ). 2 males, Chumphon Province, Pathio District, Wat Tham Khao Plu, 10°43′48″N, 99°19′15″E, 22.v.2010, P. Pimvichai leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the anterior gonopod telopodite (at) simple, far overreaching coxa (cx), apically narrow, erect, and by having the telopodital part (pt) of the posterior gonopod ending in a rounded lamella, with irregularly serrated margin (Fig. 10H).
Description
Adult males with 53 or 54 podous rings. Length ~5–6 cm, diameter ~4.2–4.5 mm. Adult females with 51–53 podous rings. Length ~5–6 cm, diameter ~4.5–4.9 mm.
Colour of living animal yellowish brown. Antennae and legs reddish brown, dorsal prozona creamy yellow, lateral side of prozona and metazona dark brown, mid dorsal metazona with a dark brown small triangular spot, collum and preanal process light brown (Fig. 13F).
Anterior gonopods (Fig. 10A, B, D, E) with high, triangular coxae, lateral margin concave at ½ of its height, apically abruptly narrowed, pointed; at base of posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite simple, far overreaching coxa, apically narrow, erected.
Posterior gonopods (Fig. 10C, F–H) very simple, with short, smooth coxal part (pcx); telopodital part (pt) very long, curving mesad, forming a deep concavity, apically rounded, with serrate margin.
Female vulvae (Fig. 10I) simple, valves prominent, the right valve larger than the left valve.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329004 (voucher code CUMZ-D00131).
Distribution
Chumphon Province, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin noun in apposition, meaning ‘tiger’ and referring to the colour pattern on living specimens (Fig. 13F).
Coxobolellus transversalis, sp. nov.
Material examined
Holotype. Male, Thailand, Nan Province, Muang District, Tham Pha Tub, 18°51′17″N, 100°44′10″E, 9.vi.2019, P. Pimvichai and T. Backeljau leg. (CUMZ).
Paratypes. Thailand: 5 females, same data as holotype (CUMZ).
Diagnosis
Differing from all other species in the genus by having the tip the of anterior gonopod coxae transversely truncated, and by having the telopodital part (pt) of the posterior gonopod fairly long compared with the coxal part (pcx).
Description
Adult male with 53 podous rings. Length ~6 cm, diameter ~4.4 mm. Adult females with 50–52 podous rings. Length ~7–8 cm, diameter ~4.0–4.9 mm.
Colour of living animal dark green.
Anterior gonopods (Fig. 11A, B, D, E) with high coxae, apically truncated, mesal margins straight, diverging, delimiting a V-shaped space between both coxae, posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite overreaching coxa, apically narrow, forming triangular process with pigmented brown node, with extremely constricted lateral margin at 2/3 of its height, forming a big triangular process.
Posterior gonopods (Fig. 11C, F) very simple, with long, smooth coxal part (pcx); telopodital part (pt) fairly long, curving mesad, flattened, concave forming a canopy, with strong transverse ridge near base, with a short spine protruding from surface near tip.
Female vulvae (Fig. 11G, H): valves prominent, of equal size.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329007 (voucher code CUMZ-D00125).
Distribution
Known only from the type locality in Nan Province, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin adjective referring to the transverse truncation of the anterior gonopod coxa.
Coxobolellus valvatus, sp. nov.
Material examined
Holotype. Male, Thailand, Chiang-Mai Province, Chiang Dao District, Wat Tham Chiang Dao, 19°23′ 37″N, 98°55′42″E, 8.i.2008, C. Sutcharit leg. (CUMZ).
Paratypes. Thailand: 1 male, 1 female, Chiang-Mai Province, Chom Thong District, Tham Borichinda, 18°30′03″N, 98°40′20″E, 7.x.2007, P. Pimvichai, S. Panha and H. Enghoff leg. (CUMZ). 1 male, 1 female, Mae Hong Son Province, Muang District, Tham Sam Ta, 19°31′23″N, 98°05′22″E, 19.vii.2008, S. Panha leg. (CUMZ).
Diagnosis
Differing from all other species in the genus by having the outer process of the anterior gonopod coxa broadly rounded, the inner process protruding higher than the outer one, and by having the telopodital part (pt) of the posterior gonopod ending in a rounded margin with a sharp spine protruding from the mesal surface near the tip. Further differing from all other species, except C. nodosus, sp. nov., by having the tip of the anterior gonopod coxa concave.
Description
Adult males with 47 or 48 podous rings. Length ~6–8 cm, diameter ~4.4–4.7 mm. Adult females with 47 or 48 podous rings. Length ~7 cm, diameter ~5.0–5.2 mm.
Colour after 11–12 years in ethanol: head, antenna and legs light brown; body rings dark brown.
Anterior gonopods (Fig. 12A, B, D, E) with high coxae, apically concave, outer process broadly rounded, inner process small, triangular; posterior surface with fairly high ridge laterally for accommodation of telopodite. Telopodite overreaching coxa, apically narrowly rounded with pigmented brown node, with constricted lateral margin at ~⅔ of its height, forming a triangular process.
Posterior gonopods (Fig. 12C, F–I) very simple, with long, smooth coxal part (pcx); telopodital part (pt) ending in a rounded margin with a sharp spine protruding from mesal surface near tip.
Female vulvae (Fig. 12J): valves prominent, of equal size; with one fairly large triangular tooth, valves fitting tightly together.
DNA barcode
The GenBank accession number of the barcode of the holotype is MT329009 (voucher code CUMZ-D00127).
Distribution
Known from Chiang-Mai and Mae Hong Son provinces, Thailand (Fig. 14).
Etymology
The specific epithet is a Latin adjective referring to the prominently toothed vulva valve.

|
|
1 Tip of anterior gonopod coxa concave or bilobed or forming a triangular process |
|
2 Tip of anterior gonopod coxa obliquely truncated (Fig. 3A, D); telopodital part (pt) of posterior gonopod short compared to coxal part (pcx) (Fig. 3C, F) |
|
3 Tip of anterior gonopod coxa forming triangular process |
|
4 Tip of anterior gonopod coxa concave, forming equal outer and inner lobes (Fig. 6A, D); telopodital part of posterior gonopod (pt) ending in a long, sharp spine, with a flattened lamella protruding from mesal surface near tip (Fig. 6C, F–H) |
|
5 Tip of anterior gonopod coxa ending in a simple triangular process |
|
6 Tip of anterior gonopod telopodite (at) forming a triangular process |
|
7 Telopodital part (pt) of posterior gonopod without a sharp, curling lamella at base (Fig. 8C, F) |
|
8 Anterior gonopod telopodite (at) far overreaching anterior gonopod coxa (cx), with narrowed tip |
|
9 Anterior gonopod telopodite (at) curving laterad (Fig. 7B, E); telopodital part (pt) of posterior gonopod laterally with serrate margin (Fig. 7C, F–H) |
Discussion
The 10 new species described here form a well supported clade in our mtDNA analysis, sister to a clade consisting of two species of Pseudospirobolellus. Unfortunately, of the other allegedly pseudospirobolellid genus, Benoitolus, we have only a COI sequence of B. birgitae, but no data for the type species, B. flavicollis. However, Coxobolellus, gen. nov. is well characterised and supported by morphological characters, notably the posterior gonopod telopodite divided into two parts, coxal part (pcx) and telopodital part (pt); coxae of the 3rd (and 4th) with extremely large, protruding process; and with the opening pore (oeg) at the mesal margin at the end of the coxal part (pcx). In other pseudospirobolellids, the 3rd leg-pair has no lobes, but lobes are found on the 5th leg-pair instead (B. flavicollis: H. Enghoff (pers. obs.) on specimens from Singapore in NHMD; B. birgitae: Hoffman (1981) [as Solaenobolellus birgitae]; B. siamensis: Attems (1936) [as Cyclothyrophorus siamensis], P. avernus: H. Enghoff (pers. obs.) on specimens from Fiji in NHMD; P. sigmoides: unknown). Moreover, the posterior gonopods of the 10 new species are clearly different from those of Benoitolus and Pseudospirobolellus. The posterior gonopod is extremely slender in Pseudospirobolellus or slender in Benoitolus and without any process, whereas in the 10 new Coxobolellus, gen. nov. species the posterior gonopod is broad and clearly divided into two parts (pcx and pt) and obviously with an opening of the efferent groove (oeg). Furthermore, the COI sequence divergence between Coxobolellus, gen. nov. and Pseudospirobolellus or Benoitolus is 20–23%. From this study, the COI sequence divergence between Coxobolellus, gen. nov and (1) pachybolid genera is 16–25% (mean: 21%), (2) trigoniulid genera is 15–21% (mean: 18%), (3) Rhinocricus parcus is 24–26% (mean: 25%), (4) Narceus annularis is 21–23% (mean: 22%), and (5) Paraspirobolus lucifugus is 23–25% (mean: 24%). The COI sequence divergence among pachybolid genera is 13–23% (mean: 19%). The COI sequence divergence between B. birgitae and pachybolid genera is 11–24% (mean: 21%). The lowest COI sequence divergence is with Litostrophus (11–18%; mean: 15%). In the order Spirostreptida, family Spirostreptidae the intergeneric COI sequence divergences are 6.83–26.81% (mean: 18.43%) (Mwabvu et al. 2015). Hence, taken together, the amount of COI sequence divergence between Coxobolellus, gen. nov. and other pseudospirobolellid, pachybolid and spirostreptid genera supports its recognition as a separate genus.
The intraspecific COI sequence divergence of Coxobolellus, gen. nov. species ranges from 0 to 5% (mean: 2%), and the interspecific divergence ranges from 6 to 15% (mean: 11%). These figures correspond well with the amounts of intra- and interspecific COI sequence divergence observed in other millipede genera. In the widespread pill millipede, Glomeris marginata (Villers, 1789) the maximum intraspecific COI sequence divergence is 5% and the interspecific divergences between G. marginata and other Glomeris species range from 12.9 to 15.9% (Reip and Wesener 2018). The intraspecific COI sequence divergence of Bavarian Diplopoda varies from 0 to 6.61% (mean: 0.82) and the mean interspecific divergence is 14.17% (Spelda et al. 2011). High intraspecific COI sequence divergences are also found in the harpagophorid genus Thyropygus Pocock, 1894 (0 to 9%), whereas interspecific divergences ranged from 5 to 18% (mean: 14%) (Pimvichai et al. 2014).
Our mtDNA analysis assigns Benoitolus birgitae to the Pachybolidae, i.e. phylogenetically very distant from Coxobolellus, gen. nov. and Pseudospirobolellus. Conversely, while the prefemoral pads on legs of male Coxobolellus, gen. nov. constitute a very unusual character, it is shared with B. birgitae, B. flavicollis and P. avernus, although in the latter two species, the pads are much less prominent (Hoffman 1981; P. Pimvichai, pers. obs.). Hence it remains to be decided whether the morphological similarity of the prefemoral pads on the legs of male Coxobolellus, gen. nov., Pseudospirobolellus and Benoitolus represents a synapomorphy contradicting our mtDNA data (which assign B. birgitae to Pachybolidae), or involves a case of homoplasy that is consistent with our mtDNA data. In a similar spirit, Enghoff et al. (2015) already expressed doubts with respect to the monophyly of the Pseudospirobolellidae. Hence further phylogenetic analyses are needed to explore the monophyly and relationships of this still poorly known family. Moreover, since this study is based on only two mtDNA gene fragments and fairly few specimens, future phylogenetic analyses would benefit from including nuclear gene markers and more specimens.
Nevertheless, this first DNA study of the Pseudospirobolellidae shows that there still is an overwhelming amount of hidden taxonomic diversity to be discovered in the south-east Asian diplopod fauna, even in supposedly species-poor families such as the Pseudospirobolellidae.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This research was funded by the Thailand Science Research and Innovation (TSRI) as a TRF Research Career Development Grant (2019–2021; RSA6280051) (to P. Pimvichai). Additional funding came from the Royal Belgian Institute of Natural Sciences (RBINS).
Acknowledgements
We thank Chirasak Sutcharit, Pongpun Prasankok and members of the Animal Systematics Research Unit, Chulalongkorn University for assistance in collecting material. We are indebted to Nesrine Akkari (NHMW) for providing specimens, to Julien Cillis (RBINS) for help with SEM photographs, to Yves Barette (RBINS) for help with gonopod photographs and to Thita Krutchuen (Chulalongkorn University) for the excellent drawings.
References
Attems, C. (1936). Diplopoda of India. Memoirs of the Indian Museum 11, 133–323.Attems, C. (1953). Myriopoden von Indochina. Expedition von Dr C. Dawydoff (1938–1939). Mémoires du Muséum National d’Histoire Naturelle. Série A, Zoologie 5, 133–230.
Carl, J. (1912). Die Diplopoden-Fauna von Celebes. Revue Suisse de Zoologie 20, 73–206.
| Die Diplopoden-Fauna von Celebes.Crossref | GoogleScholarGoogle Scholar |
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29, 1969–1973.
| Bayesian phylogenetics with BEAUti and the BEAST 1.7.Crossref | GoogleScholarGoogle Scholar | 22367748PubMed |
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797.
| MUSCLE: multiple sequence alignment with high accuracy and high throughput.Crossref | GoogleScholarGoogle Scholar | 15034147PubMed |
Enghoff, H., Golovatch, S., Short, M., Stoev, P., and Wesener, T. (2015). Diplopoda – taxonomic overview. In ‘The Myriapoda 2. Treatise on Zoology – Anatomy, Taxonomy, Biology’. (Ed. A. Minelli.) pp. 363–453. (Brill: Leiden, Netherlands.)
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3, 294–299.
| 7881515PubMed |
Fujisawa, T., and Barraclough, T. G. (2013). Delimiting species using single-locus data and the generalized mixed yule coalescent approach: a revised method and evaluation on simulated data sets. Systematic Biology 62, 707–724.
| Delimiting species using single-locus data and the generalized mixed yule coalescent approach: a revised method and evaluation on simulated data sets.Crossref | GoogleScholarGoogle Scholar | 23681854PubMed |
Hebert, P. D. N., Cywinska, A., Ball, S. L., and DeWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London – B. Biological Sciences 270, 313–321.
| Biological identifications through DNA barcodes.Crossref | GoogleScholarGoogle Scholar |
Hillis, D., and Bull, J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42, 182–192.
| An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar |
Hoffman, R. L. (1981). Studies on spiroboloid millipeds. XIV. Notes on the family Pseudospirobolellidae, and the description of a new genus and species from Thailand. Steenstrupia 7, 181–190.
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755.
| MRBAYES: Bayesian inference of phylogeny.Crossref | GoogleScholarGoogle Scholar | 11524383PubMed |
Jeekel, C. A. W. (2001). A bibliographic catalogue of the Spirobolida of the Oriental and Australian Regions (Diplopoda). Myriapod Memoranda 4, 3–104.
Kekkonen, M., Mutanen, M., Kaila, L., Nieminen, M., and Hebert, P. D. N. (2015). Delineating species with DNA barcodes: a case of taxon dependent method performance in moths. PLoS One 10, e0122481.
| Delineating species with DNA barcodes: a case of taxon dependent method performance in moths.Crossref | GoogleScholarGoogle Scholar | 25849083PubMed |
Kessing, B., Croom, H., Martin, A., McIntosh, C., McMillan, W. O., and Palumbi, S. (2004). PCR primers. In ‘The Simple Fool’s Guide to PCR’. Version 1.0, pp. 17–18. (University of Hawaii, Department of Zoology: Honolulu, HI, USA.)
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547–1549.
| MEGA X: molecular evolutionary genetics analysis across computing platforms.Crossref | GoogleScholarGoogle Scholar | 29722887PubMed |
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34, 772–773.
| PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar | 28013191PubMed |
Likhitrakarn, N., Golovatch, S. I., and Panha, S. (2011). Revision of the Southeast Asian millipede genus Orthomorpha Bollman, 1893, with the proposal of a new genus (Diplopoda, Polydesmida, Paradoxosomatidae). ZooKeys 131, 1–161.
| Revision of the Southeast Asian millipede genus Orthomorpha Bollman, 1893, with the proposal of a new genus (Diplopoda, Polydesmida, Paradoxosomatidae).Crossref | GoogleScholarGoogle Scholar |
Mauriès, J.-P. (1980). Contributions à l’étude de la faune terrestre des îles granitiques de l’archipel des Séchelles (Mission P.L.G. Benoit – J.J. Van Mol 1972). Myriapoda – Diplopoda. Revue de Zoologie Africaine 94, 138–168.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In ‘Proceedings of the Gateway Computing Environments Workshop (GCE)’, 14 November 2010, New Orleans, LA, USA. INSPEC Accession Number: 11705685, pp. 1–8. (IEEE.)
Mwabvu, T., Lamb, J., Slotow, R., Hamer, R. M., and Barraclough, D. (2015). Do cytochrome c oxidase 1 gene sequences differentiate species of spirostreptid millipedes (Diplopoda: Spirostreptida: Spirostreptidae)? African Invertebrates 56, 651–661.
| Do cytochrome c oxidase 1 gene sequences differentiate species of spirostreptid millipedes (Diplopoda: Spirostreptida: Spirostreptidae)?Crossref | GoogleScholarGoogle Scholar |
Pimvichai, P., Enghoff, H., and Panha, S. (2014). Molecular phylogeny of the Thyropygus allevatus group of giant millipedes and some closely related groups. Molecular Phylogenetics and Evolution 71, 170–183.
| Molecular phylogeny of the Thyropygus allevatus group of giant millipedes and some closely related groups.Crossref | GoogleScholarGoogle Scholar | 24269316PubMed |
Pimvichai, P., Enghoff, H., and Panha, S. (2016). A revision of the Thyropygus allevatus group. Part V: Nine new species of the extended opinatus subgroup, based on morphological and DNA sequence data (Diplopoda: Spirostreptida: Harpagophoridae). European Journal of Taxonomy 199, 1–37.
| A revision of the Thyropygus allevatus group. Part V: Nine new species of the extended opinatus subgroup, based on morphological and DNA sequence data (Diplopoda: Spirostreptida: Harpagophoridae).Crossref | GoogleScholarGoogle Scholar |
Pimvichai, P., Enghoff, H., Panha, S., and Backeljau, T. (2018). Morphological and mitochondrial DNA data reshuffle the taxonomy of the genera Atopochetus Attems, Litostrophus Chamberlin and Tonkinbolus Verhoeff (Diplopoda: Spirobolida: Pachybolidae), with descriptions of nine new species. Invertebrate Systematics 32, 159–195.
| Morphological and mitochondrial DNA data reshuffle the taxonomy of the genera Atopochetus Attems, Litostrophus Chamberlin and Tonkinbolus Verhoeff (Diplopoda: Spirobolida: Pachybolidae), with descriptions of nine new species.Crossref | GoogleScholarGoogle Scholar |
Pitz, K. M., and Sierwald, P. (2010). Phylogeny of the millipede order Spirobolida (Arthropoda: Diplopoda: Helminthomorpha). Cladistics 26, 497–525.
| Phylogeny of the millipede order Spirobolida (Arthropoda: Diplopoda: Helminthomorpha).Crossref | GoogleScholarGoogle Scholar |
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21, 1864–1877.
| ABGD, Automatic Barcode Gap Discovery for primary species delimitation.Crossref | GoogleScholarGoogle Scholar | 21883587PubMed |
Reip, H. S., and Wesener, T. (2018). Intraspecific variation and phylogeography of the millipede model organism, the black pill millipede Glomeris marginata (Villers, 1789) (Diplopoda, Glomerida, Glomeridae). ZooKeys 741, 93–131.
| Intraspecific variation and phylogeography of the millipede model organism, the black pill millipede Glomeris marginata (Villers, 1789) (Diplopoda, Glomerida, Glomeridae).Crossref | GoogleScholarGoogle Scholar | 30872937PubMed |
Shiels, D. R., Hurlbut, D. L., Lichtenwald, S. K., and Monfils, A. K. (2014). Monophyly and phylogeny of Schoenoplectus and Schoenoplectiella (Cyperaceae): evidence from chloroplast and nuclear DNA sequences. Systematic Botany 39, 132–144.
| Monophyly and phylogeny of Schoenoplectus and Schoenoplectiella (Cyperaceae): evidence from chloroplast and nuclear DNA sequences.Crossref | GoogleScholarGoogle Scholar |
Spelda, J., Reip, H. S., Oliveira–Biener, U., and Melzer, R. R. (2011). Barcoding Fauna Bavarica: Myriapoda – a contribution to DNA sequence-based identifications of centipedes and millipedes (Chilopoda, Diplopoda). ZooKeys 156, 123–139.
| Barcoding Fauna Bavarica: Myriapoda – a contribution to DNA sequence-based identifications of centipedes and millipedes (Chilopoda, Diplopoda).Crossref | GoogleScholarGoogle Scholar |
Srisonchai, R., Enghoff, H., Likhitrakarn, N., and Panha, S. (2018a). A revision of dragon millipedes I: genus Desmoxytes Chamberlin, 1923, with the description of eight new species (Diplopoda, Polydesmida, Paradoxosomatidae). ZooKeys 761, 1–177.
| A revision of dragon millipedes I: genus Desmoxytes Chamberlin, 1923, with the description of eight new species (Diplopoda, Polydesmida, Paradoxosomatidae).Crossref | GoogleScholarGoogle Scholar |
Srisonchai, R., Likhitrakarn, N., Enghoff, H., and Panha, S. (2018b). A revision of dragon millipede IV: the new genus Spinaxytes, with the description of nine new species (Diplopoda, Polydesmida, Paradoxosomatidae). ZooKeys 797, 19–69.
| A revision of dragon millipede IV: the new genus Spinaxytes, with the description of nine new species (Diplopoda, Polydesmida, Paradoxosomatidae).Crossref | GoogleScholarGoogle Scholar |
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
| RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Crossref | GoogleScholarGoogle Scholar | 24451623PubMed |
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729.
| MEGA6: molecular evolutionary genetics analysis version 6.0.Crossref | GoogleScholarGoogle Scholar | 24132122PubMed |
Xia, X. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Molecular Biology and Evolution 35, 1550–1552.
| DAMBE7: new and improved tools for data analysis in molecular biology and evolution.Crossref | GoogleScholarGoogle Scholar | 29669107PubMed |