SSR allelic diversity in relation to morphological traits and resistance to grain mould in sorghum
Rajan Sharma A B , S. P. Deshpande A , S. Senthilvel A , V. P. Rao A , V. Rajaram A , C. T. Hash A and R. P. Thakur AA International Crops Research Institute for the Semi-Arid Tropics, Patancheru, Hyderabad 502 324, Andhra Pradesh, India.
B Corresponding author. Email: r.sharma@cgiar.org
Crop and Pasture Science 61(3) 230-240 https://doi.org/10.1071/CP09192
Submitted: 3 July 2009 Accepted: 22 December 2009 Published: 9 March 2010
Abstract
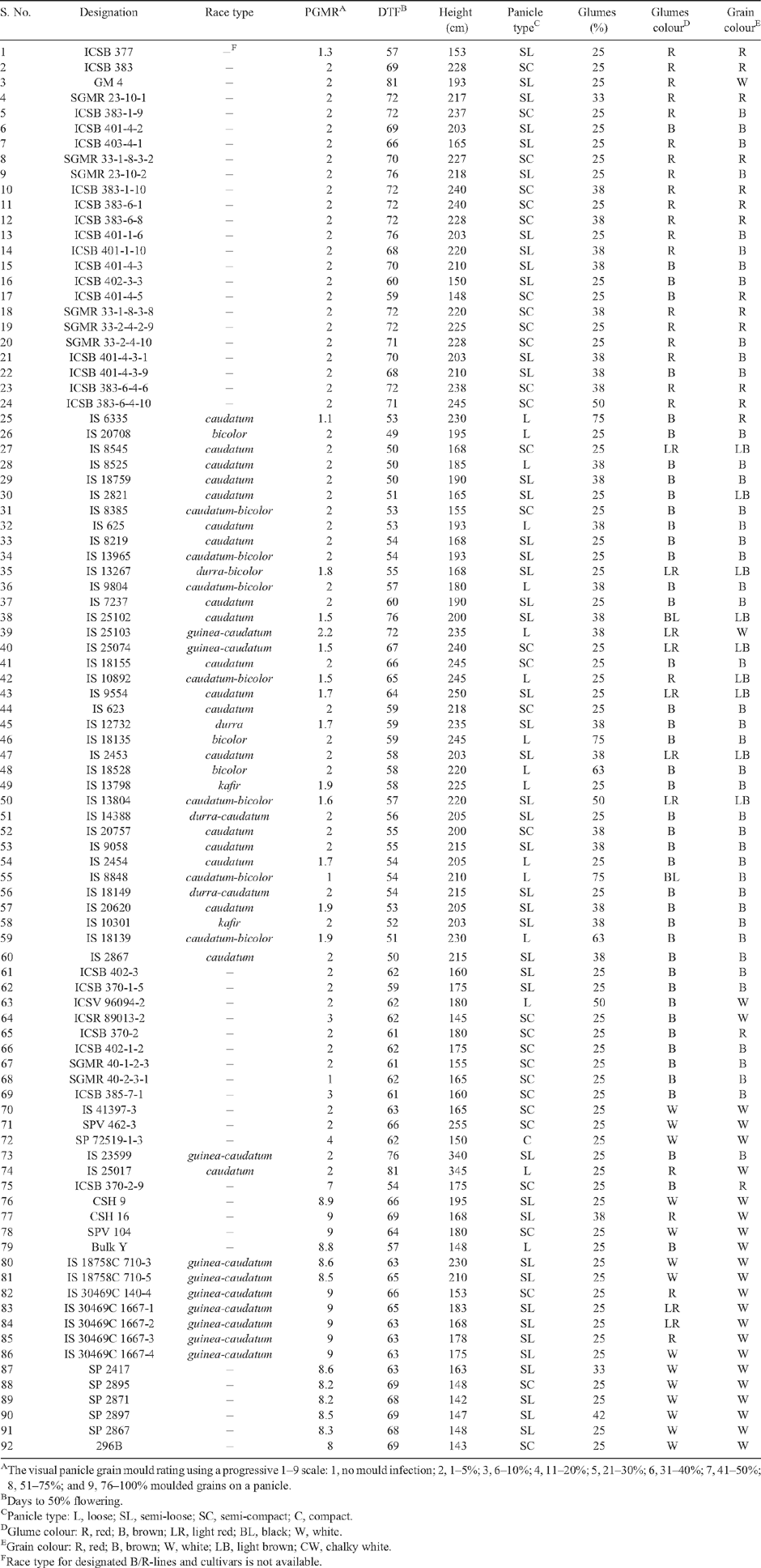
Allelic variation at 46 simple sequence repeat (SSR) marker loci well distributed across the sorghum genome was used to assess genetic diversity among 92 sorghum lines, 74 resistant and 18 susceptible to grain mould. Of the 46 SSR markers, 44 were polymorphic, with the number of alleles ranging from 2 to 20 with an average of 7.55 alleles per locus. Genetic diversity among the sorghum lines was high as indicated by polymorphic information content (PIC) and gene diversity values. PIC values of polymorphic SSR markers ranged from 0.16 to 0.90, with an average of 0.54. Gene diversity among the sorghum lines varied from 0.16 to 0.91, with an average score of 0.58 per SSR marker. AMOVA indicated that 12% of the total variation observed among the sorghum lines was accounted for between grain mould resistant and susceptible types. Diversity based on six morphological traits and grain mould scores indicated major roles of panicle type and glumes coverage, followed by grain colour, in clustering of the lines. Seven grain mould resistant/susceptible pairs with dissimilarity indices >0.50, but with similar flowering time, plant height, and panicle type/inflorescence within each pair, were selected for use in developing recombinant inbred line mapping populations to identify genomic regions (and quantitative trait loci) associated with sorghum grain mould resistance.
Additional keywords: diversity, grain mould, sorghum, SSR.
Acknowledgments
The research was supported by The Sehgal Foundation Endowment Fund instituted at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, Andhra Pradesh, India.
Agrama HA,
Widle GE,
Reese JC,
Campbell LR, Tuinstra MR
(2002) Genetic mapping of QTLs associated with green bug resistance and tolerance in Sorghum bicolor. Theoretical and Applied Genetics 104, 1373–1378.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Audilakshmi S,
Stenhouse JW,
Reddy TP, Prasad MVR
(1999) Grain mold resistance and associated characters of sorghum genotypes. Euphytica 107, 91–103.
| Crossref | GoogleScholarGoogle Scholar |

Audilakshmi S,
Stenhouse JW, Reddy TP
(2005) Genetic analysis of grain mold resistance in white seed sorghum genotypes. Euphytica 145, 95–101.
| Crossref | GoogleScholarGoogle Scholar |

Bhattramakki D,
Dong J,
Chhabra AK, Hart GE
(2000) An integrated SSR and RFLP linkage map of Sorghum bicolor (L.) Moench. Genome 43, 988–1002.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Botstein D,
White RL,
Skolnick M, Davis RW
(1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32, 314–331.
|
CAS |
PubMed |

Brown SM,
Hopkins MS,
Mitchell SE,
Senior ML,
Wang TY,
Duncan RR,
Gonzalez-Candelas F, Kresovich S
(1996) Multiple methods for the identification of polymorphic simple sequence repeats (SSRs) in sorghum [Sorghum bicolor (L.) Moench]. Theoretical and Applied Genetics 93, 190–198.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Casa AM,
Mitchell SE,
Hamblin MT,
Sun H,
Bowers JE,
Paterson AH,
Aquadro CF, Kresovich S
(2005) Diversity and selection in sorghum: Simultaneous analyses using simple sequence repeats (SSRs). Theoretical and Applied Genetics 111, 23–30.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Esele JP,
Frederiksen RA, Miller FR
(1993) The association of genes controlling caryopsis traits with grain mold resistance in sorghum. Phytopathology 83, 490–495.
| Crossref | GoogleScholarGoogle Scholar |

Excoffier L,
Laval G, Schneider S
(2005) Arlequin ver. 3.0: An integrated software package for population genetic data analysis. Evolutionary Bioinformatics Online 1, 47–50.
|
CAS |
PubMed |

Feltus FA,
Hart GE,
Schertz KF,
Casa AM,
Kresovich S,
Abraham P,
Klein PE,
Brown PJ, Paterson AH
(2006) Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theoretical and Applied Genetics 112, 1295–1305.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Folkertsma RT,
Rattunde HFW,
Chandra S,
Soma Raju G, Hash CT
(2005) The pattern of genetic diversity of guinea-race Sorghum bicolor (L.) Moench land races as revealed with SSR markers. Theoretical and Applied Genetics 111, 399–409.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Geleta N,
Labuschagne MT, Viljoen CD
(2006) Genetic diversity analysis in sorghum germplasm as estimated by AFLP, SSR and morpho-agronomical markers. Biodiversity and Conservation 15, 3251–3265.
| Crossref | GoogleScholarGoogle Scholar |

Ghebru B,
Schmidt RJ, Bennetzen JL
(2002) Genetic diversity of Eritrean sorghum landraces assessed with simple sequence repeat (SSR) markers. Theoretical and Applied Genetics 105, 229–236.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Gower JC, Legendre P
(1986) Metric and Euclidean properties of dissimilarity coefficients. Journal of Classification 3, 5–48.
| Crossref | GoogleScholarGoogle Scholar |

Harris HB, Burns RE
(1973) Relationship between tannin content of sorghum grain and preharvest seed molding. Agronomy Journal 65, 957–959.
|
CAS |

Harris K,
Subudhi PK,
Borrell A,
Jordan D,
Rosenow D,
Nguyen H,
Klein P,
Klein R, Mullet J
(2007) Sorghum stay-green QTL individually reduce post-flowering drought-induced leaf senescence. Journal of Experimental Botany 58, 327–338.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Idury MI, Cardon LR
(1997) A simple method for automated allele binning in microsatellite markers. Genome Research 7, 1104–1109.
|
CAS |
PubMed |

Kim JS,
Klein PE,
Klein RR,
Price HJ,
Mullet JE, Stelly DM
(2005) Chromosome identification and nomenclature of Sorghum bicolor. Genetics 169, 1169–1173.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Klein PE,
Klein RR,
Cartinhour SW,
Ulanch PE,
Dong J,
Obert JA,
Morshige DT,
Schlueter SD,
Childs KL,
Ale M, Mullet JE
(2000) A high-throughput AFLP-based method for constructing integrated genetic and physical maps: Progress toward a sorghum genome map. Genome Research 10, 789–807.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Klein RR,
Rodriguez-Herrera R,
Schlueter JA,
Klein PE,
Yu ZH, Rooney WL
(2001) Identification of genomic regions that affect grain mold incidence and other traits of agronomic importance in sorghum. Theoretical and Applied Genetics 102, 307–319.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Kong L,
Dong J, Hart GE
(2000) Characteristics, linkage map positions, and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple sequence repeats (SSRs). Theoretical and Applied Genetics 101, 438–448.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Liu K, Muse SV
(2005) PowerMarker: Integrated analysis environment for genetic molecular data. Bioinformatics 21, 2128–2129.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mace ES,
Buhariwalla HK, Crouch JH
(2003) A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Molecular Biology Reporter 21, 459–460.
| Crossref | GoogleScholarGoogle Scholar |

Mace ES,
Rami J-F,
Bouchet S,
Klein PE,
Klein RR,
Kilian A,
Wenzl P,
Xia L,
Halloran K, Jordan DR
(2009) A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers. BMC Plant Biology 9, 13.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Magalhaes JV,
Garvin DF,
Wang Y,
Sorrells ME,
Klein PE,
Schaffert RE,
Li L, Kochian LV
(2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae. Genetics 167, 1905–1914.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mansuetus SBA,
Frederiksen RA,
Waniska RD,
Odvody GN, Craig J
(1990) The role of glumes of sorghum in resistance to grain mold. Phytopathology [Abstract] 80, 1069.

Mantel NA
(1967) The detection of disease clustering and a generalized regression approach. Cancer Research 27, 209–220.
|
CAS |
PubMed |

Menkir A,
Goldsbrough P, Ejeta G
(1997) RAPD based assessment of genetic diversity in cultivated races of sorghum. Crop Science 37, 564–569.
|
CAS |

Menz MA,
Klein RR,
Mullet JE,
Obert JA,
Unruh NC, Klein PE
(2002) A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Molecular Biology 48, 483–499.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Mittal M, Boora KS
(2006) Molecular tagging of gene conferring leaf blight resistance using microsatellites in sorghum [Sorghum bicolor (L.) Moench]. Indian Journal of Experimental Biology 43, 462–466.

Nagy ER,
Lee T-C,
Ramakrishna W,
Xu Z,
Klein PE,
San Miguel P,
Cheng C-P,
Li J,
Devos KM,
Schertz K,
Dunkle L, Bennetzen JL
(2007) Fine mapping of the Pc locus of Sorghum bicolor, a gene controlling the reaction to a fungal pathogen and its host-selective toxin. Theoretical and Applied Genetics 114, 961–970.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Oh B-J,
Frederiksen RA, Magill CW
(1994) Identification of molecular markers linked to head smut resistance gene (SHS) in sorghum by RFLP and RAPD analysis. Phytopathology 84, 830–833.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Oh B-J,
Frederiksen RA, Magill CW
(1996) Identification of RFLP markers linked to a gene for downy mildew resistance (Sdm) in sorghum. Canadian Journal of Botany 74, 315–317.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Paterson AH,
Bowers JE,
Bruggmann R,
Dubchak I,
Grimwood J,
Gundlach H,
Haberer G,
Hellsten U,
Mitros T,
Poliakov A,
Schmutz J,
Spannagl M,
Tang H,
Wang X,
Wicker T,
Bharti AK,
Chapman J,
Feltus FA,
Gowik U,
Grigoriev IV,
Lyons E,
Maher CA,
Martis M,
Narechania A,
Otillar RP,
Penning BW,
Salamov AA,
Wang Y,
Zhang L,
Carpita NC,
Freeling M,
Gingle AR,
Hash CT,
Keller B,
Klein P,
Kresovich S,
McCann MC,
Ming R,
Peterson DG,
Mehboob-ur-Rahman
,
Ware D,
Westhoff P,
Mayer KFX,
Messing J, Rokhsar DS
(2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ramu P,
Kassahun B,
Senthilvel S,
Kumar CA,
Jayashree B,
Folkertsma RT,
Reddy LA,
Kuruvinashetti MS,
Haussmann BIG, Hash CT
(2009) Exploiting rice–sorghum synteny for targeted development of EST-SSRs to enrich the sorghum genetic linkage map. Theoretical and Applied Genetics 119, 1193–1204.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Ramu P,
Deshpande SP,
Senthilvel S,
Jayashree B,
Billot C,
Deu M,
Ananda Reddy L, Hash CT
(2010) In-silico mapping of important genes and markers available in public domain for efficient sorghum breeding. Molecular Breeding , (In press).

Reddy BVS,
Sharma HC,
Thakur RP, Ramesh S
(2006) Characterization of ICRISAT-bred sorghum hybrid parents (set I). International Sorghum and Millet Newsletter 47, 1–21.

Saitou N, Nei M
(1987) The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425.
|
CAS |
PubMed |

Schloss SJ,
Mitchell SE,
White GM,
Kukatla R,
Bowers JE,
Paterson AH, Kresovich S
(2002) Characterization of RFLP clone sequences for gene discovery and SSR development in Sorghum bicolor (L). Moench. Theoretical and Applied Genetics 105, 912–920.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Smith JSC,
Kresovich S,
Hopkins MS,
Mitchell SE,
Dean RE,
Woodman WL,
Lee M, Porter K
(2000) Genetic diversity among elite sorghum inbred lines assessed with simple sequence repeats. Crop Science 40, 226–232.
|
CAS |

Taramino G,
Tarchini R,
Ferrario S,
Lee M, Pe’ ME
(1997) Characterization and mapping of simple sequence repeats (SSRs) in Sorghum bicolor. Theoretical and Applied Genetics 95, 66–72.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Thakur RP,
Rao VP,
Navi SS,
Garud TB,
Agarkar GD, Bharati B
(2003) Sorghum grain mold: Variability in fungal complex. International Sorghum and Millet Newsletter 4, 104–108.

Ward JH
(1963) Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association 58, 236–244.
| Crossref | GoogleScholarGoogle Scholar |

Weir BS, Hill WG
(2002) Estimating F-statistics. Annual Review of Genetics 36, 721–750.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Williams RJ, Rao KN
(1981) A review of sorghum grain moulds. Tropical Pest Management 27, 200–211.
| Crossref | GoogleScholarGoogle Scholar |

Wright S
(1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19, 395–420.
| Crossref | GoogleScholarGoogle Scholar |

|


