On the nose: validating a novel, non-invasive method to identify individual koalas using unique nose patterns
Janine Duffy A , Tori Stragliotto B and Valentina S. A. Mella B *
B *
A
B
Abstract
Non-invasive identification of individual animals in wild populations can occur in species with unique coat patterns (e.g. zebras, giraffes, jaguars). However, identification in other species often relies on capture-mark–recapture techniques, involving physical handling of animals. Identification of individual koalas (Phascolarctos cinereus) is difficult and has so far relied mostly on invasive methods such as ear tagging, microchipping and/or collaring, which require capture. The validation of a non-invasive method to identify koalas could improve monitoring of individuals in the wild, allowing targeting of specific koalas in disease and survival studies, reducing the need to capture individuals.
This study describes a novel effective method to identify koalas from their nose markings, specifically using the unpigmented pattern of the nose to determine unique features of individuals.
Photographs of koalas from different populations in Victoria and New South Wales (NSW), Australia, were examined in the study. Nose patterns were traced from photographs and matched through visual assessment if they were thought to belong to the same individual. Differences in identification success between datasets from different populations and the effect of sex on match success were evaluated statistically. For the NSW koalas, the effect of lighting conditions and photographic angle were also assessed.
Overall identification success was 89.7% (range 87.1–91.8%) and was not affected by any of the variables tested, demonstrating that nose patterns can be used reliably to identify individual koalas.
The proposed non-invasive method is simple, yet accurate and stable over time, hence it offers a vital tool for monitoring endangered koalas whilst minimising human interference.
Pattern-based recognition of koalas is cost-effective, reduces stress on the animals, has the potential to improve data collection and allows involvement of citizen scientists in monitoring of populations or individuals.
Keywords: biometric features, citizen science, individual identification, mark-recapture, nose markings, pattern-based identification, Phascolarctos cinereus, photographic identification.
Introduction
Long-term identification of individual animals in the wild is important for the study of wildlife behaviour and for conservation and management research, in particular for threatened and endangered species (Clutton-Brock and Sheldon 2010). However, methods such as banding, tagging or branding are not always cost-effective, are considered invasive and can affect natural occurring behaviours (Wilson and McMahon 2006; Walker et al. 2012). Non-invasive identification methods can be used in wildlife species when individuals present unique biometric features, such as fin marks and notches in cetaceans (Wuersig and Jefferson 1990; Gowans and Whitehead 2001), plumage markings in birds (Bretagnolle et al. 1994; Usher et al. 2016), ear shape in elephants (Ardovini et al. 2008; Bedetti et al. 2020) and facial wrinkles in rhinoceros (Patton and Campbell 2011).
Pigment pattern-based identification is commonly used in species presenting unique markings of the coat or body, such as large cats like jaguars, Panthera onca (Nipko et al. 2020), cheetahs, Acinonyx jubatus (Caro 1994), tigers, Panthera tigris (Karanth et al. 2006; Hiby et al. 2009), zebras (Petersen 1972; Prinsloo et al. 2021), giraffes, Giraffa camdopardulis (Foster 1966; Shorrocks and Croft 2009), but also birds (Sherley et al. 2010), reptiles (Knox et al. 2013; Yamamoto et al. 2022; Gould et al. 2024), and anuran (Lama et al. 2011; Lassnig et al. 2024).
Photographic identification and recognition of free-ranging individual animals from biometric features have been used extensively in both marine and terrestrial habitats (Karczmarski et al. 2022). The methods used for individual identification vary from manual (e.g. conducted by one or more observers and checked against an image library or identification matrix; Wuersig and Jefferson 1990; Tye et al. 2015; Möcklinghoff et al. 2018), to computer aided or fully automated (e.g. pattern recognition softwares and deep learning algorithms; Kelly 2001; Van Tienhoven et al. 2007; Anderson et al. 2010; Nipko et al. 2020; Das et al. 2021).
Individual identification of animals with little or no distinct body markings is more complex (Güthlin et al. 2014). However, pattern identification has also been used in domestic bovine and canine species, using unique grooves and ridges of the nose of individuals as morphometric features (Kumar and Singh 2017; Kumar et al. 2018; Chui and Karczmarski 2022). These identification methods have been validated to be accurate (90–98%) through statistical analysis, advanced deep learning algorithms (point feature algorithm extraction in unconstrained conditions), and deep belief networks (feedforward neural network analysis for image recognition) (Kumar et al. 2018; Choi et al. 2021). Therefore, identification of individuals using unique phenotypical and/or morphological features represents an important tool for recognition also in species where individuals are hard to discern.
The koala, Phascolarctos cinereus, is an endangered native Australian marsupial (Environment Protection And Biodiversity Conservation Act 1999) that is considered a flagship species for Australia biodiversity and is integral to the tourism industry (Hundloe and Hamilton 1997). Conservation efforts for the koala have recently increased driven by attention from the public and Government to prevent the species from becoming extinct (Department of Agriculture Water and the Environment 2022). Part of the conservation work implemented is to monitor koala populations through surveys or individual koala health through clinical signs of disease (Griffith 2010) but individual koalas are difficult to identify. Long-term individual monitoring for health and survival relies on microchipping and tagging techniques considered invasive and involving capture and restraint of animals, which are expensive and time consuming, as they require experienced catching personnel and permits (Radford et al. 2006; Madani et al. 2020).
Currently, there are no validated non-invasive methods for koalas that can identify individuals accurately. However, recent literature has reported that citizen scientists use nose patterns and other features such as markings around the chin to identify individual koalas, making the species suitable for visual identification techniques (Danaher et al. 2023). We describe here a non-invasive method developed by Janine Duffy from the Koala Clancy Foundation (https://www.koalaclancyfoundation.org.au/) in Victoria that follows pattern recognition theory in koalas, using nose patterns as identifying features for individuals. We used photographs to investigate the success rate of nose pattern identification for wild koalas from four different areas in Australia, in Victoria and in New South Wales.
Materials and methods
Defining a nose pattern
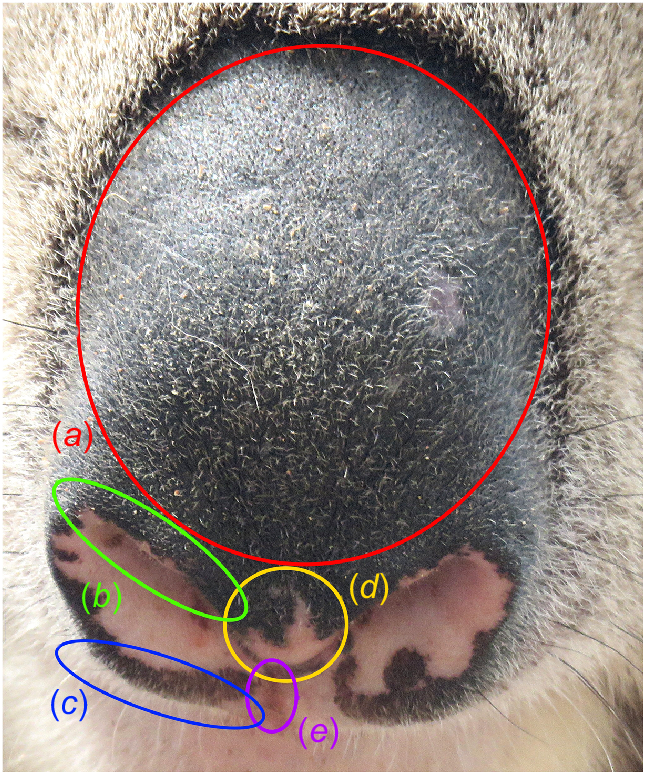
Koalas have large, prominent, bare-skin noses that are mostly pigmented black, but the inside of the nostrils is usually unpigmented (Kratzing 1984). The nose pattern is defined here as the unpigmented shape inside and just outside the nostrils, including along the nostril ridge, philtrum, nostril tip and to the upper lip (Fig. 1). For individual recognition, the outline of the nose pattern is assessed for major feature points, including projections, notches and spots of unpigmented skin. Unpigmented shapes on the planum (Fig. 1) of the nose may also be present, but these are usually a result of age or trauma, and although nose scars have been used for identification in other species (Gilkinson et al. 2007), these features are not considered for identification in koalas.
Longitudinal photo-ID of free-ranging koalas
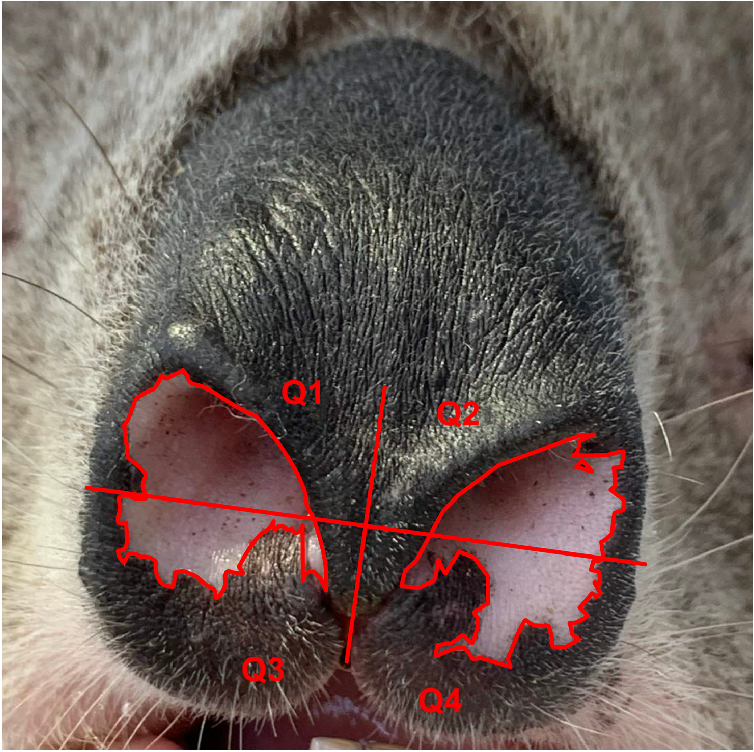
Field photographs of individual koalas in the You Yangs Regional Park (YYRP) and Brisbane Ranges in Victoria, Australia were taken opportunistically (range 2–1200 times) from August 2001 to November 2023. Each time an individual koala was encountered, it was photographed with a hand-held digital camera (Canon EOS 600D, Canon Powershot SX 60, Olympus C-730 or similar) from the ground, from a distance of at least 10 m. Photographs were taken focused on the nose area, using optical or digital zoom. Nose patterns were later outlined (Fig. 2) using a graphics editor and design software such as Adobe Illustrator (Adobe Inc 2024) using quadrants to identify feature points in specific areas of the nose. A database of koala noses was then created. For each new photograph, the nose pattern of the koala was checked against already existing images in the database and assigned to a particular individual if previous images matched the current one.
Photo-ID – koala-ID validation
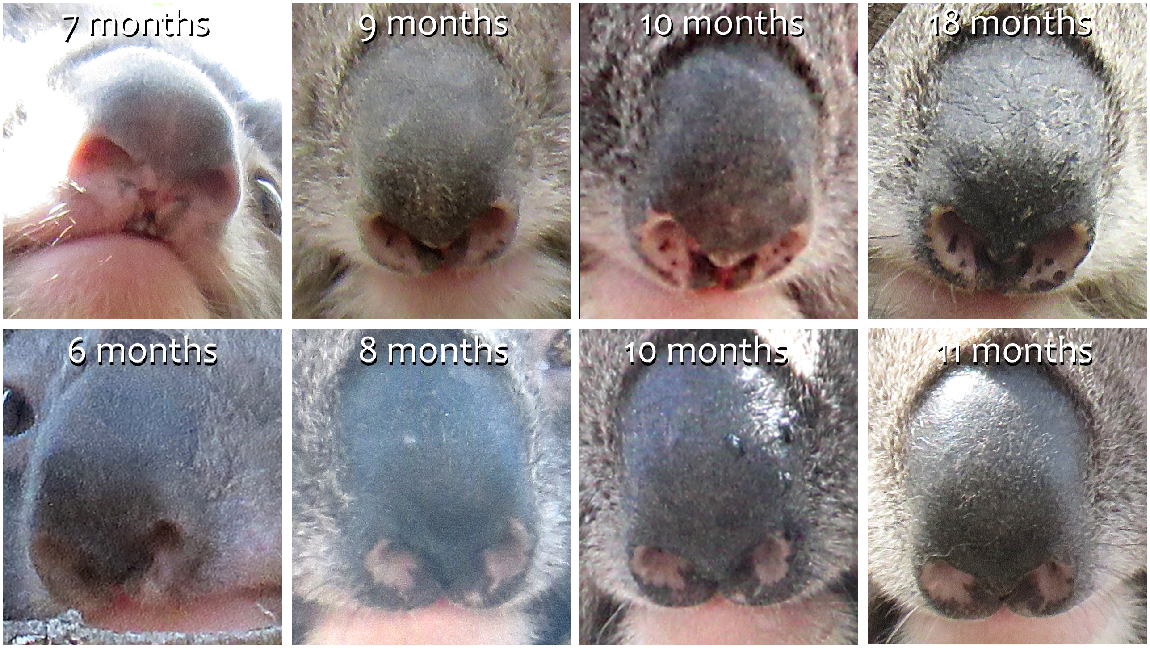
We randomly selected photographs (N = 113) of 51 adult koalas (22 males and 29 females) in the database to establish if wild free-ranging individuals in trees could be identified by their nose patterns over time (range 3–13 years). The photographs (N = 77) of 15 koalas (six males and nine females) photographed at pouch emergence (min. 8 months) and at least once again in adulthood (max. 9 years) were used to establish if nose patterns change with development. Nose pattern of very young joeys (6 months) was not discernible, so could not be used for identification purposes.
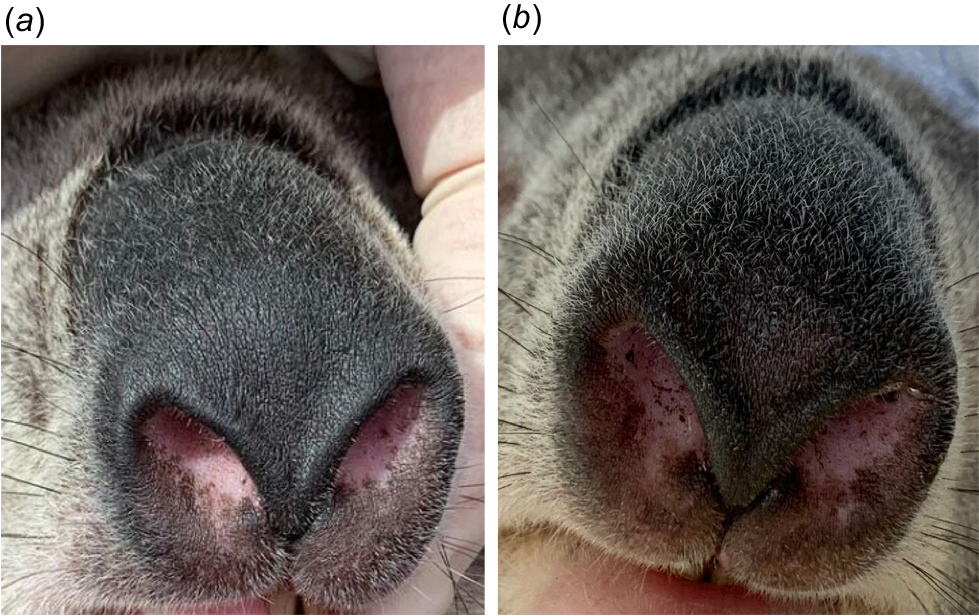
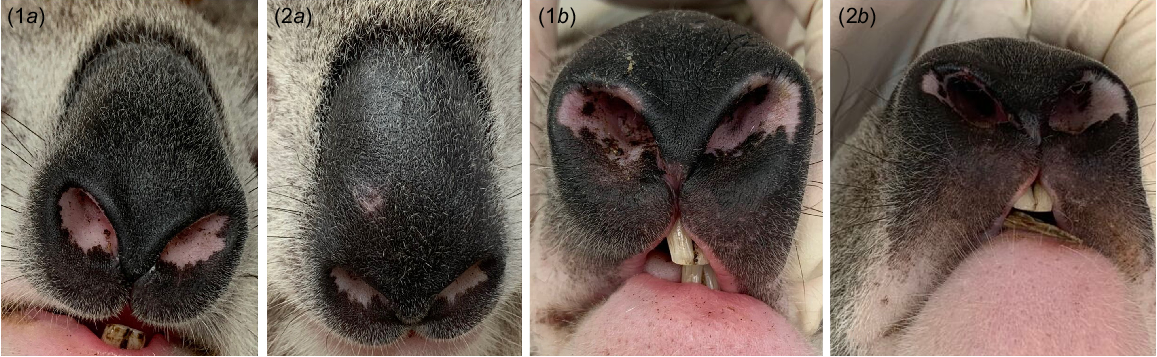
For geographical validation of the methodology, we also used photographs (n = 176) of individual koalas (N = 86, 40 males and 46 females) used for research between November 2022 and July 2023 from two distinct koala populations, in the Southern Highlands (N = 99) and Gunnedah (N = 77) in New South Wales (NSW), Australia. These koalas were captured, brought to the ground, sedated and given a microchip as part of a different study (Simpson et al. 2023). Hence, individual identification for these koalas could be verified undoubtedly. A digital camera (Canon EOS 600D) was used at close range to photograph the same koalas on at least two separate occasions (minimum 3 months apart). These photographs varied in light conditions (dark = 63, light = 113; Fig. 3) and angle (frontal = 59, angled = 117; Fig. 4).
Example of light (a) and dark (b) photographs of the nose of the same individual koala (Phascolarctos cinereus) from New South Wales, Australia.
Example of frontal (1a and 2a) and angled (1b and 2b) photos of two different individual (1 and 2) koalas (Phascolarctos cinereus) from New South Wales, Australia.
At least two people (range 2–5), blinded to the identities of photographed koalas, conducted photo-ID matching for each of the three datasets available: (1) wild free-ranging adult koalas in trees (Victoria), (2) wild free-ranging joeys in trees (Victoria) and (3) wild captured koalas on the ground (NSW). Datasets were randomly allocated to observers (at least two per observer). All observers were naive to koala identification and were trained by an expert observer. The outlines of the nose patterns of the individuals in the pictures were compared to identify possible matches. Finally, photos were checked against database pictures (for free-ranging individuals) or microchip details (for captured individuals). Correct photo-ID matches were assigned a 1 and unsuccessful matches a 0.
Data analysis
A generalised linear mixed effect model (GLMM) with a binomial distribution was used to examine whether successful photo matches to the koala ID (binary, 0 = fail, 1 = success) were affected by the dataset used (i.e. Victoria = koalas in trees, New South Wales = koalas on the ground) and sex (male or female) in R (R Development Core Team 2023), using the ‘lme4’ package and the ‘glmer’ function (Searle et al. 1980). Koala ID and observer ID were the random factors. For the koalas in trees in Victoria, we used another GLMM to check if identification was affected by time in months (to establish that both adult koalas and joeys are identifiable over time). For the wild captured koalas in New South Wales, we checked the effect of photographic variables (i.e. photo lighting and photo angle) on successful identification using a separate GLMM, as in other species success varies with these (Sripian et al. 2015; Theimer et al. 2017).
Results
Longitudinal photo-ID of free-ranging koalas
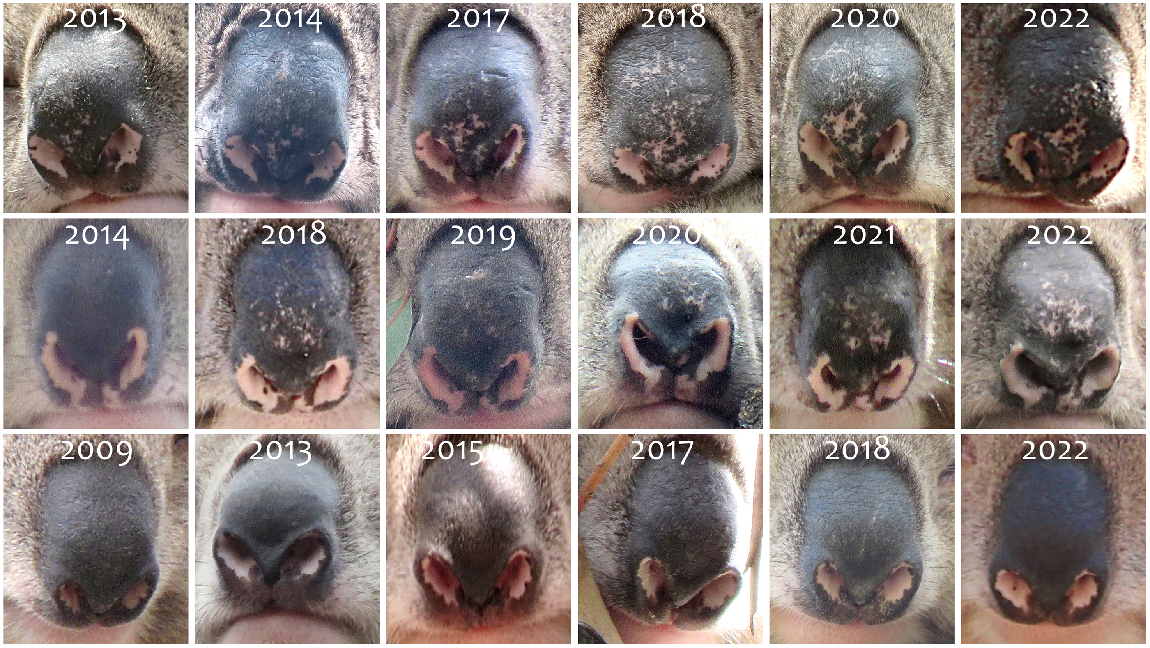
156 koalas (75 females, 78 males and 3 unknown) were included in the nose patterns database between August 2001 and November 2023 in the YYRP and Brisbane Ranges in Victoria, Australia. Most individual koalas were photographed and identified over long-time frames (i.e. 51% were identified repeatedly for a minimum of 3 years or more; Fig. 5).
Photo-ID – koala-ID validation
Overall identification success was 89.7%. For wild free-ranging adult koalas in trees (Fig. 6) identification success was 87.2% (range 86.7–87.6%), while for dependent joeys (Fig. 7) it was 90.3% (range 83.1–97.4%). For wild captured koalas photographed on the ground identification success was 91.7% (range 87.5–94.9-%). There was no significant difference in identification success between datasets (F = 0.63, P = 0.535) and no effect of sex (F = 2.93, P = 0.087) on success rate. For wild free-ranging koalas in trees (both adult and joeys) identification success did not change over time (F = 0.63, P = 0.535). For wild captured koalas, angle (F = 1.24; P = 0.213) and lighting (F = 0.05; P = 0.996) of the photograph did not influence identification success.
Discussion
We described and tested the efficacy of a novel non-invasive identification method for koalas utilising photos of nose patterns. We found individual variability in koala nose markings in four distinct koala populations in Victoria and NSW, Australia and demonstrated that the use of koala nose patterns for identification purposes is a simple and feasible method to distinguish between individual koalas in different locations, independent of sex, lighting and angle of observation. Nose patterns are stable over time but do not appear to be fully formed on pouch emergence. This method of koala recognition could be used widely in research, rehabilitation sector and citizen science when a non-invasive method is preferred, or funding is not available for microchipping and ear tagging, and as a backup to other methods.
89.7% of koala nose pattern photographs were matched successfully to individual identity. Identification of individuals by biometric features may vary in different populations and locations in pachyderms (Stein et al. 2010; Patton and Campbell 2011; Bedetti et al. 2020). However, nose patterns differences in the koalas in our study provided enough individual variation that location did not affect identification success. Our results from four populations in Victoria and NSW indicate that the koala nostril pattern identification method could be applied to other koala populations in Australia.
Nose patterns may change over time with exposure to the environment and due to injury and scarring, and this may reduce re-identification success over long periods as in other species (Patton and Campbell 2011; Bedetti et al. 2020). However, the nose patterns of koalas used in this study did not discernibly change over time as all koalas were identifiable over the years even if minor changes due to scarring increased with ageing. Our results indicate that koalas can be identified by their nose pattern from approximately 8 months old for their lifespan, despite changes to pigmentation with ageing. This is similar in the critically endangered African forest elephant (Loxodonta cyclotis) where individuals were identified with 85.2% accuracy over 15 years (Bodesheim et al. 2022). As koala nose patterns demonstrate substantial stability over time, nose marking can be considered a reliable biometric feature for the species. To note, in this study one koala in the New South Wales dataset was recorded with two different microchip numbers within the original study by Simpson et al. (2023), as the first microchip had to be replaced due to malfunction, resulting in the same individual having two different microchip IDs on record. However, this individual was able to be identified correctly by its nose markings and the dataset corrected. Hence, koala nose patterns can also be used in conjunction with microchipping and ear tagging to confirm individual identity in case of replaced identifiers, thus eliminating errors associated with tag loss or microchip malfunction in capture-mark–recapture events.
Individuals of different sexes may vary in their level of identifiability. In a study on sea otters, Enhydra lutris, sex affected sighting frequency of individuals identified by nose scars because males had more scars than females and were therefore more likely to be identifiable (Gilkinson et al. 2007). However, sex did not influence identification success of nose patterns of koalas, indicating that nose markings are reliable identification features for both male and female individuals. Observation angle and lighting also did not affect identification success. Previous studies on other species such as the African penguin (Sherley et al. 2010), the Asiatic black bear, Ursus thibetanus (Sripian et al. 2015) and several ungulates (Ness et al. 2022) have noted that lighting, contrast and image quality can pose a challenge for the identification of individuals using photographs. However, these did not affect nose pattern identification success in our study. The photos we used for this analysis were taken at close range while individuals were being handled and are of higher quality than those obtained from the ground. However, our analysis demonstrated that there was no significant difference in identification success from photos taken from a distance while koalas were in tree or at close range while koalas were on the ground. Regardless of factors that can affect image quality, the major features that allow identification of individuals are evident and recognisable in all of the photos.
The photographic outline drawing method that we used in this study to record nostril differences and identify koalas, follows the traditional approach of tracing the image and creating a bank of photographic data in which new photos can be compared (Foster 1966; Petersen 1972). The simplicity of this method of identification opens up new avenues to apply the technique to valuable research projects and opportunities to validate the method further in other populations both in the wild and in captivity. Furthermore, the development of an automated pattern recognition software for koala nose markings to improve efficiency of individual identification would be extremely valuable, as demonstrated in other species (Anderson et al. 2010; Kumar et al. 2018). Deep learning algorithms used in software identification have been estimated to save the equivalent of greater than 17,000 h of human identification efforts with 96.6% accuracy (Norouzzadeh et al. 2018).
Invasive methods of animal identification, such as tagging and collaring, often raise significant animal welfare concerns. While these methods can be necessary to achieve certain research objectives, they can also cause stress, injury, or behavioural changes (Wilson and McMahon 2006). Consequently, there has been increasing scrutiny and ethical debate surrounding the use of such invasive techniques (Beausoleil et al. 2018), particularly when dealing with threatened species, such as the koala. As artificial intelligence (AI) technology continues to advance, it is expected to play a greater role in reducing the impact of research on wildlife. AI-driven tools, such as non-invasive photographic identification and automated monitoring systems, offer promising alternatives that minimise the need for physical interventions, thus enhancing the welfare of the animals studied (Norouzzadeh et al. 2018).
The nose pattern identification method for koalas could be routinely used by researchers, rehabilitators and veterinarians to identify specific individuals and for monitoring after release in the wild. For example, in field transect surveys for population monitoring there is the risk of recounting the same individuals (Ellis et al. 2002), but the nose identification method could help reduce this risk. In the rehabilitation and veterinary sector this proposed non-invasive method could be used for post-release monitoring which is notoriously difficult due to lack of funding (Guy et al. 2013). The method could also be used for koalas returned to the wild after treatment for chlamydiosis, allowing monitoring of recurring ocular and urogenital clinical signs of disease (Robbins et al. 2018).
Conclusions
This study demonstrates that koala nose patterns can be used as a unique biometric recognition feature for koala identification, making it extremely valuable for research. The method we described is non-invasive and simple, yet accurate and stable over time, and can be readily applied for behavioural and health monitoring of free-ranging individuals. Our results provide a baseline for future research to use koala nose biometric features to develop automated efficient analysis. Researchers should consider photographing individual koala noses for identification purposes since the pattern remains consistent enough over time to allow long-term identification, and that microchips and ear tags have the potential to fail or fall off.
Data availability
The data used to generate the results in the paper will be shared on the Sydney eScholarship Repository https://ses.library.usyd.edu.au.
Acknowledgements
We would like to thank Koala Clancy Foundation staff for providing support for the project and many of the nose photographs. We also thank all the volunteers who worked on the photo identification and matching including Emer Cahill, Josh Hall-Johnston, Jack Downey and Maria Costantino.
References
Adobe Inc (2024) Adobe Illustrator. Available at https://adobe.com/products/illustrator
Anderson CJR, Da Vitoria Lobo N, Roth JD, Waterman JM (2010) Computer-aided photo-identification system with an application to polar bears based on whisker spot patterns. Journal of Mammalogy 91, 1350-1359.
| Crossref | Google Scholar |
Ardovini A, Cinque L, Sangineto E (2008) Identifying elephant photos by multi-curve matching. Pattern Recognition 41, 1867-1877.
| Crossref | Google Scholar |
Beausoleil NJ, Mellor DJ, Baker L, Baker SE, Bellio M, Clarke AS, Dale A, Garlick S, Jones B, Harvey A, Pitcher BJ, Sherwen S, Stockin KA, Zito S (2018) “Feelings and Fitness” not “Feelings or Fitness”–The Raison d’être of conservation welfare, which aligns conservation and animal welfare objectives. Frontiers in Veterinary Science 5, 296.
| Crossref | Google Scholar |
Bedetti A, Greyling C, Paul B, Blondeau J, Clark A, Malin H, Horne J, Makukule R, Wilmot J, Eggeling T, Kern J, Henley M (2020) System for Elephant Ear-pattern Knowledge (SEEK) to identify individual African elephants. Pachyderm 61, 63-77.
| Google Scholar |
Bodesheim P, Blunk J, Körschens M, Brust C-A, Käding C, Denzler J (2022) Pre-trained models are not enough: active and lifelong learning is important for long-term visual monitoring of mammals in biodiversity research – individual identification and attribute prediction with image features from deep neural networks and decoupled decision models applied to elephants and great apes. Mammalian Biology 102, 875-897.
| Crossref | Google Scholar |
Bretagnolle V, Thibault J-C, Dominici J-M (1994) Field identification of individual ospreys using head marking pattern. The Journal of Wildlife Management 58, 175-178.
| Crossref | Google Scholar |
Choi HI, Lee Y, Shin H, Lee S, Choi SS, Han CY, Kwon S-H (2021) The formation and invariance of canine nose pattern of beagle dogs from early puppy to young adult periods. Animals 11, 2664.
| Crossref | Google Scholar |
Chui SYS, Karczmarski L (2022) Everyone matters: identification with facial wrinkles allows more accurate inference of elephant social dynamics. Mammalian Biology 102, 645-666.
| Crossref | Google Scholar |
Clutton-Brock T, Sheldon BC (2010) Individuals and populations: the role of long-term, individual-based studies of animals in ecology and evolutionary biology. Trends in Ecology & Evolution 25, 562-573.
| Crossref | Google Scholar | PubMed |
Danaher M, Schlagloth R, Hewson M, Geddes C (2023) One person and a camera: a relatively non-intrusive approach to Koala citizen science. Australian Zoologist 43, 52-66.
| Crossref | Google Scholar |
Das A, Sowmya S, Sinha S, Chandru S (2021) Identification of a zebra based on its stripes through pattern recognition. In ‘2021 International Conference on Design Innovations for 3Cs Compute Communicate Control (ICDI3C)’, Bangalore, India. pp. 120–122. (IEEE) doi:10.1109/ICDI3C53598.2021.00033
Ellis WAH, Melzer A, Carrick FN, Hasegawa M (2002) Tree use, diet and home range of the koala (Phascolarctos cinereus) at Blair Athol, central Queensland. Wildlife Research 29, 303-311.
| Crossref | Google Scholar |
Environment Protection And Biodiversity Conservation Act (1999) Commonwealth of Australia. Available at https://www.legislation.gov.au/Series/C2004A00485
Foster JB (1966) The giraffe of nairobi national park: home range, sex ratios, the herd, and food. African Journal of Ecology 4, 139-148.
| Crossref | Google Scholar |
Gilkinson AK, Pearson HC, Weltz F, Davis RW (2007) Photo-identification of sea otters using nose scars. The Journal of Wildlife Management 71, 2045-2051.
| Crossref | Google Scholar |
Gould J, Beranek C, Madani G (2024) Dragon detectives: citizen science confirms photo-ID as an effective tool for monitoring an endangered reptile. Wildlife Research 51, WR23036.
| Crossref | Google Scholar |
Gowans S, Whitehead H (2001) Photographic identification of northern bottlenose whales (Hyperoodon ampullatus): sources of heterogeneity from natural marks. Marine Mammal Science 17, 76-93.
| Crossref | Google Scholar |
Güthlin D, Storch I, Küchenhoff H (2014) Is it possible to individually identify red foxes from photographs? Wildlife Society Bulletin 38, 205-210.
| Crossref | Google Scholar |
Guy AJ, Curnoe D, Banks PB (2013) A survey of current mammal rehabilitation and release practices. Biodiversity and Conservation 22, 825-837.
| Crossref | Google Scholar |
Hiby L, Lovell P, Patil N, Kumar NS, Gopalaswamy AM, Karanth KU (2009) A tiger cannot change its stripes: using a three-dimensional model to match images of living tigers and tiger skins. Biology Letters 5, 383-386.
| Crossref | Google Scholar | PubMed |
Karanth KU, Nichols JD, Kumar NS, Hines JE (2006) Assessing tiger population dynamics using photographic capture–recapture sampling. Ecology 87, 2925-2937.
| Crossref | Google Scholar | PubMed |
Karczmarski L, Chan SCY, Rubenstein DI, Chui SYS, Cameron EZ (2022) Individual identification and photographic techniques in mammalian ecological and behavioural research—Part 1: methods and concepts. Mammalian Biology 102, 545-549.
| Crossref | Google Scholar |
Kelly MJ (2001) Computer-aided photograph matching in studies using individual identification: an example from Serengeti cheetahs. Journal of Mammalogy 82, 440-449.
| Crossref | Google Scholar |
Knox CD, Cree A, Seddon PJ (2013) Accurate identification of individual geckos (Naultinus gemmeus) through dorsal pattern differentiation. New Zealand Journal of Ecology 37, 60-66.
| Google Scholar |
Kratzing JE (1984) The anatomy and histology of the nasal cavity of the koala (Phascolarctos cinereus). Journal of Anatomy 138, 55-65.
| Google Scholar | PubMed |
Kumar S, Singh SK (2017) Automatic identification of cattle using muzzle point pattern: a hybrid feature extraction and classification paradigm. Multimedia Tools and Applications 76, 26551-26580.
| Crossref | Google Scholar |
Kumar S, Pandey A, Sai Ram Satwik K, Kumar S, Singh SK, Singh AK, Mohan A (2018) Deep learning framework for recognition of cattle using muzzle point image pattern. Measurement 116, 1-17.
| Crossref | Google Scholar |
Lama FD, Rocha MD, Andrade MÂ, Nascimento LB (2011) The use of photography to identify individual tree frogs by their natural marks. South American Journal of Herpetology 6, 198-204.
| Crossref | Google Scholar |
Lassnig N, Guasch-Martínez S, Pinya S (2024) The individual color pattern on the back of Bufotes viridis balearicus (Boettger, 1880) allows individual photo-identification recognition for population studies. Canadian Journal of Zoology 102, 393-402.
| Crossref | Google Scholar |
Madani GF, Ashman KR, Mella VSA, Whisson DA (2020) A review of the ‘noose and flag’ method to capture free-ranging koalas. Australian Mammalogy 42, 341-348.
| Crossref | Google Scholar |
Möcklinghoff L, Schuchmann K-L, Marques MI (2018) New non-invasive photo-identification technique for free-ranging giant anteaters (Myrmecophaga tridactyla) facilitates urgently needed field studies. Journal of Natural History 52, 2397-2411.
| Crossref | Google Scholar |
Ness IFG, Jung TS, Schmiegelow FKA (2022) Evaluating likelihood-based photogrammetry for individual recognition of four species of northern ungulates. Mammalian Biology 102, 701-718.
| Crossref | Google Scholar |
Nipko RB, Holcombe BE, Kelly MJ (2020) Identifying individual jaguars and ocelots via pattern-recognition software: comparing HotSpotter and Wild-Id. Wildlife Society Bulletin 44, 424-433.
| Crossref | Google Scholar |
Norouzzadeh MS, Nguyen A, Kosmala M, Swanson A, Palmer MS, Packer C, Clune J (2018) Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proceedings of the National Academy of Sciences 115, E5716-E5725.
| Crossref | Google Scholar |
Patton FJ, Campbell PE (2011) Using eye and profile wrinkles to identify individual white rhinos. Pachyderm 50, 84-86.
| Google Scholar |
Petersen JCB (1972) An identification system for zebra (Equus burchelli, Gray. African Journal of Ecology 10, 59-63.
| Crossref | Google Scholar |
Prinsloo ND, Postma M, de Bruyn PJN (2021) How unique is unique? Quantifying geometric differences in stripe patterns of Cape mountain zebra, Equus zebra zebra (Perissodactyla: Equidae). Zoological Journal of the Linnean Society 191, 612-625.
| Crossref | Google Scholar |
Radford SL, Mckee J, Goldingay RL, Kavanagh RP (2006) The protocols for koala research using radio-collars: a review based on its application in a tall coastal forest in New South Wales and the implications for future research projects. Australian Mammalogy 28, 187-200.
| Crossref | Google Scholar |
R Development Core Team (2023) ‘R: a language and environment for statistical computing. Version 4.3.1.’ (R Foundation for Statistical Computing: Vienna, Austria) Available at https://www.R-project.org/
Robbins A, Loader J, Timms P, Hanger J (2018) Optimising the short and long-term clinical outcomes for koalas (Phascolarctos cinereus) during treatment for chlamydial infection and disease. PLoS ONE 13, e0209679.
| Crossref | Google Scholar | PubMed |
Searle SR, Speed FM, Milliken GA (1980) Population marginal means in the linear model: an alternative to least squares means. The American Statistician 34, 216-221.
| Crossref | Google Scholar |
Sherley RB, Burghardt T, Barham PJ, Campbell N, Cuthill IC (2010) Spotting the difference: towards fully-automated population monitoring of African penguins Spheniscus demersus. Endangered Species Research 11, 101-111.
| Crossref | Google Scholar |
Shorrocks B, Croft DP (2009) Necks and networks: a preliminary study of population structure in the reticulated giraffe (Giraffa camelopardalis reticulata de Winston). African Journal of Ecology 47, 374-381.
| Crossref | Google Scholar |
Simpson SJ, Higgins DP, Timms P, Mella VSA, Crowther MS, Fernandez CM, McArthur C, Phillips S, Krockenberger MB (2023) Efficacy of a synthetic peptide Chlamydia pecorum major outer membrane protein vaccine in a wild koala (Phascolarctos cinereus) population. Scientific Reports 13, 15087.
| Crossref | Google Scholar | PubMed |
Stein AB, Erckie B, Fuller TK, Marker L (2010) Camera-trapping as a method for monitoring rhino populations within the Waterberg Plateau Park, Namibia. Pachyderm 48, 67-70.
| Google Scholar |
Theimer TC, Ray DT, Bergman DL (2017) Camera angle and photographic identification of individual striped skunks. Wildlife Society Bulletin 41, 146-150.
| Crossref | Google Scholar |
Tye CA, Greene DU, Giuliano WM, Mccleery RA (2015) Using camera-trap photographs to identify individual fox squirrels (Sciurus niger) in the Southeastern United States. Wildlife Society Bulletin 39, 645-650.
| Crossref | Google Scholar |
Usher KM, Groom C, Saunders DA (2016) Identification of individual Carnaby’s Cockatoos Calyptorhynchus latirostris from distinctive plumage markings. Australian Zoologist 38, 62-82.
| Crossref | Google Scholar |
Van Tienhoven AM, Den Hartog JE, Reijns RA, Peddemors VM (2007) A computer-aided program for pattern-matching of natural marks on the spotted raggedtooth shark Carcharias taurus. Journal of Applied Ecology 44, 273-280.
| Crossref | Google Scholar |
Walker KA, Trites AW, Haulena M, Weary DM (2012) A review of the effects of different marking and tagging techniques on marine mammals. Wildlife Research 39, 15-30.
| Crossref | Google Scholar |
Wilson RP, McMahon CR (2006) Measuring devices on wild animals: what constitutes acceptable practice? Frontiers in Ecology and the Environment 4, 147-154.
| Crossref | Google Scholar |
Yamamoto T, Nakamura T, Toda M (2022) Verification of natural marking for individual identification using a duplex marking approach in Ijima’s sea snakes, Emydocephalus ijimae (Reptilia: Elapidae). Zoological Studies 61, e75.
| Crossref | Google Scholar |