Assembly and comparative analyses of the mitochondrial genome of Castanospermum australe (Papilionoideae, Leguminosae)
Rong Zhang A B , Jian-Jun Jin A , Michael J. Moore C and Ting-Shuang Yi A D
A D
A Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Lanhei Road, Kunming, 650201, PR China.
B University of Chinese Academy of Sciences, Yuquan Road, Beijing, 100049, PR China.
C Department of Biology, Oberlin College, College Street, Oberlin, OH 44074, USA.
D Corresponding author. Email: tingshuangyi@mail.kib.ac.cn
Australian Systematic Botany 32(6) 484-494 https://doi.org/10.1071/SB19014
Submitted: 11 February 2019 Accepted: 5 April 2019 Published: 1 October 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Plant mitochondrial genomes are often difficult to assemble because of frequent recombination mediated by repeats. Only a few mitochondrial genomes have been characterised in subfamily Papilionoideae of Leguminosae. Here, we report the complete mitochondrial genome of Castanospermum australe A.Cunn. & C.Fraser, an important medicinal and ornamental species in the Aldinoid clade of Papilionoideae. By mapping paired-end reads, seven hypothetical subgenomic conformations were rejected and two hypothetical complete isometric mitochondrial genome conformations that differed by a 64-kb inversion were strongly supported. Quantitative assessment of repeat-spanning read pairs showed a major conformation (MC1) and a minor conformation (MC2). The complete mitochondrial genome of C. australe was, thus, generated as 542 079 bp in length, with a high depth of coverage (~389.7×). Annotation of this mitochondrial genome yielded 58 genes encoding 37 proteins, 18 tRNAs and three rRNAs, as well as 17 introns and three medium-sized repeats (133, 119 and 114 bp). Comparison of 10 mitochondrial genomes from Papilionoideae demonstrated significant variation in genome size, structure, gene content and RNA editing sites. In addition, mitochondrial genes were shown to be potentially useful in resolving the deep relationships of Papilionoideae.
Additional keywords: genome skimming, legume, mitochondrion, Moreton Bay chestnut, phylogeny, read-pair mapping.
Introduction
In contrast to the explosion of plastid and animal mitochondrial genome sequencing over the past 15 years, the pace of plant mitochondrial genome sequencing has been relatively slow owing to its extreme structural variation (Smith and Keeling 2015). Likewise, fewer studies in plant phylogenetics utilise mitochondrial genes because of their low substitution rate (Hiesel et al. 1994). More than 100 angiosperm mitochondrial genomes (http://www.ncbi.nlm.nih.gov/genome/organelle/, accessed 10 March 2019) have been sequenced, and show remarkable variation in genome size, content and structure (Knoop et al. 2011; Mower et al. 2012a; Guo et al. 2016). Sequenced angiosperm mitochondrial genomes range from 66 kb in Viscum scurruloideum Barlow (Skippington et al. 2015) to more than 11.3 Mb in Silene L. (Sloan et al. 2012). The almost 200-fold difference of mitochondrial genome size has been primarily attributed to the expansion or contraction of intergenic sequences. In angiosperms, pseudogene formation in mitochondrial genes as a result of functional transfer to the nucleus is frequent (Adams et al. 2002). The structure of most angiosperm mitochondrial genomes is extremely variable. This results from frequent homologous recombination mediated by repeats (Maréchal and Brisson 2010; Gualberto and Newton 2017), which can make proper assembly of mitochondrial genomes very challenging with short-read sequences. Recent studies have leveraged the increasing availability of paired-end libraries to complete assemblies of mitochondrial genomes (e.g. Naito et al. 2013; Guo et al. 2016, 2017; Yu et al. 2018). Here, we employ these techniques to sequence and assemble the mitochondrial genome of Castanospermum australe, an economically important legume species.
Only a few Leguminosae mitochondrial genomes have been reported, most included within subfamily Papilionoideae, which is the largest subfamily of Leguminosae, with ~503 genera and 14 000 described species (Legume Phylogeny Working Group 2017). Mitochondrial genome sizes in Papilionoideae range from 217 kb in Medicago truncatula Gaertn. (Bi et al. 2016) to 588 kb in Vicia faba L. (Negruk 2013). These mitochondrial genomes share 30 protein-coding genes and three rRNA genes (Shi et al. 2018), as well as the loss of three ribosomal protein genes (rps2, rps11 and rps13). Several other protein-coding genes (including cox2, rpl2, rps1, rps19, sdh3 and sdh4) are functional in some Papilionoideae mitochondrial genomes, but pseudogenised or lost in others, with a functional copy having been transferred to the nuclear genome (Palmer et al. 2000; Shi et al. 2018). Papilionoideae mitochondrial genomes also demonstrate great variation in the number and length of repeat sequences. Some mitochondrial genomes have high numbers of long repeats, such as in Glycine max (L.) Merr., which has many large repeats that have mediated recombination to produce an enriched molecular pool of 760 circles (Chang et al. 2013). Likewise, Vicia faba has 15 repeats >1 kb in length, with the longest repeat of 66 kb. In contrast, a few legume mitochondrial genomes have only short repeats of <1 kb in length, such as Vigna radiata (L.) R.Wilczek, which has only six repeats of >100 bp (Alverson et al. 2011a).
Castanospermum australe, the Moreton Bay chestnut or black bean, is the only species of the genus Castanospermum A.Cunn ex Hook. This species is native to north-eastern Australia, but has been introduced as an ornamental tree into India, South Africa and warm temperate regions of North America. The seeds of C. australe yield castanospermine, which is a major toxic alkaloidal constituent that inhibits replication of the human immunodeficiency virus (HIV; Walker et al. 1987) and other retroviruses (Sunkara et al. 1987), and reduces tumour growth in mice (Ostrander et al. 1988). Castanospermum australe belongs to the Aldinoid clade, one of the early branching lineages of Papilionoideae (Legume Phylogeny Working Group 2013). Mitochondrial genomes of a few major clades of Papilionoideae have been sequenced, including the Cladrastis clade, the Genistoid clade, the Hologalegina clade, and the Millettioid clade. However, no mitochondrial genome of the Aldinoid clade has been reported.
The purpose of the present study was to sequence and assemble the mitochondrial genome of C. australe and to compare it to other Papilionoideae mitochondrial genomes to help clarify the diversification of mitochondrial genomes in this subfamily. Given the challenges of assembling mitochondrial genomes, it is important to verify alternative conformations mediated by repeats. To accomplish this, we employed a computational approach that was developed to use read-pair counts to estimate alternative conformations (e.g. Alverson et al. 2011b; Mower et al. 2012a). Across Papilionoideae, we undertook comparative analyses of mitochondrial genome size, gene content, structural variation and RNA editing patterns. We also tested the utility of mitochondrial genomes in resolving the deep relationships of the subfamily.
Materials and methods
Genome sequencing and assembly
Fresh leaves of C. australe were collected from the Brisbane Botanic Garden, Queensland, Australia. The voucher specimen (Yi14471) was deposited in the herbarium of the Kunming Institute of Botany, Chinese Academy of Sciences (KUN). No specific permits were required for the collection of the material. Total DNA was isolated from silica gel-dried leaves of C. australe by using a modified CTAB protocol described in Yang et al. (2014). The DNA sample was sequenced from a paired-end library with an insert size of 350 bp at Beijing Genomics Institute (Shenzhen, China) on an Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA), generating ~8.6 million 150-bp paired-end reads. Reads were deposited in the NCBI Sequence Read Archive (Accession SRR8648141).
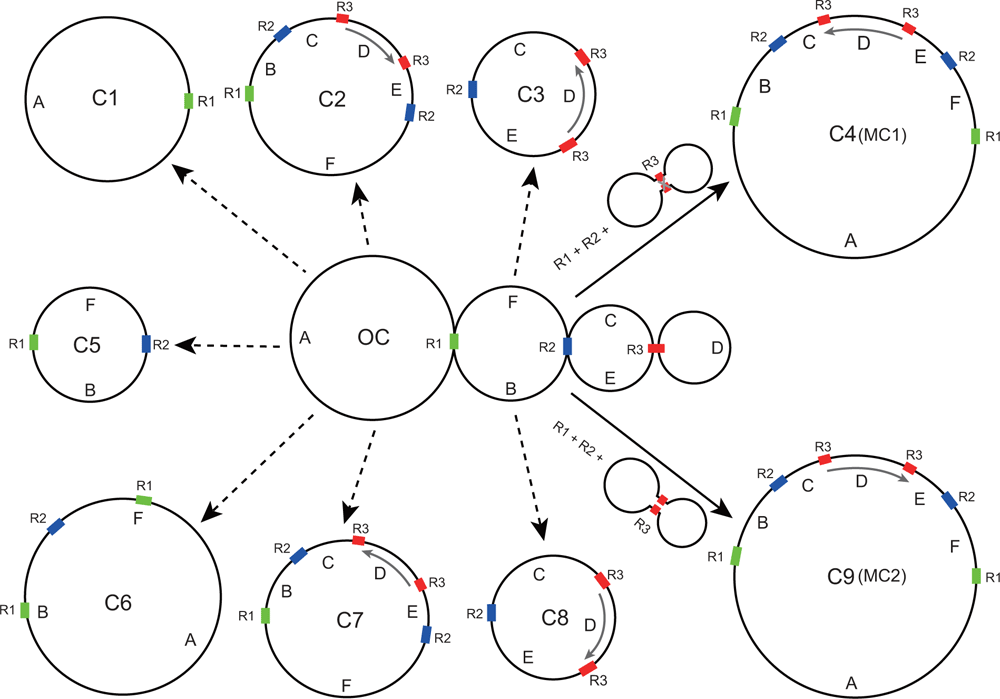
Genome-sequence data were assembled de novo using Linux-OS SPAdes genome assembler (ver. 3.10.1, see http://cab.spbu.ru/software/spades/; Bankevich et al. 2012), with the following parameters: careful; k-mer values: 75, 89, 105, 113, 121; all other parameters were set to defaults. The use of iterative k-mer lengths in SPAdes leverages the full potential of paired-end reads. Assembled contigs were then filtered and binned into mitochondrial and plastid contigs by using the Python script ‘slim_fastg_by_blast’ (see https://github.com/Kinggerm/GetOrganelle, accessed August 2016). To avoid assembly errors at regions of shared homology among the different genomes, we carefully rechecked and deleted the plastid and nuclear DNA contigs, as described preciously (Mower et al. 2012a). The final hypothesised conformations (Fig. 1, more details in Results and discussion) were manually exported using Bandage Ubuntu dynamic (ver. 8.0, see http://rrwick.github.io/Bandage/; Wick et al. 2015).
Mitochondrial genome assembly verification
To evaluate sequence-assembly quality and accuracy, paired-end reads were mapped to the assembled mitochondrial genomes using Bowtie2 (ver. 2.3.2, see http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; Langmead and Salzberg 2012) with default parameters. Read maps were visually inspected in Geneious (ver. 9.1.14, see https://www.geneious.com/; Kearse et al. 2012). Seven subgenomic and two complete genomic conformations were hypothesised on the basis of three medium-size repeats (Fig. 1). Sequencing reads were set as pairs and mapped to these nine hypothetical structures. The following parameters were used for Bowtie2: end-to-end alignment type, medium sensitivity assembly, and a maximum insert size of 600 bp.
Annotation of C. australe mitochondrial genome
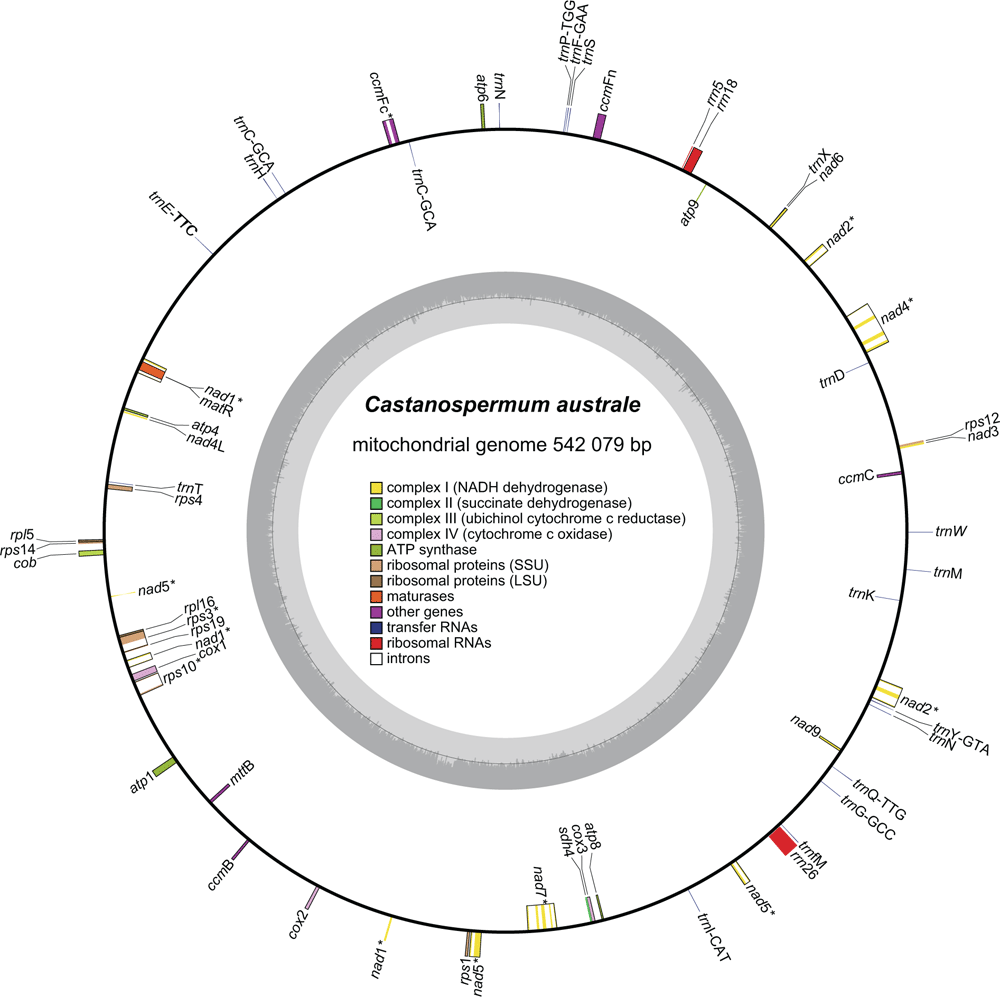
Genes, introns and tRNAs were annotated manually by using the previously published Vigna radiata (NC_015121), Glycine max (NC_020455) and Arabidopsis thaliana (L.) Heynh. (NC_001284) mitochondrial genomes as query sequences in Geneious, employing a basic local alignment search tool (BLAST)-like algorithm to search for annotations with ≥80% sequence similarity. Start and stop codons and intron splice sites were visually inspected and adjusted as necessary. A mitochondrial genome map was generated using OGDraw (see https://chlorobox.mpimp-golm.mpg.de/OGDraw.html; Lohse et al. 2013). The final annotated genome was deposited in GenBank (Accession MK426679).
Synteny comparisons of mitochondrial genomes
The mitochondrial genome structure of C. australe was compared with all available Papilionoideae mitochondrial genomes, including those of Ammopiptanthus mongolicus (Kom.) S.H.Cheng (MF683210), Glycine max (NC_020455), Lotus japonicus (Regel) Larsen (NC_016743), Medicago truncatula (NC_029641), Pongamia pinnata (L.) Pierre (NC_016742), Styphnolobium japonicum (L.) Schott. (MG757109), Vicia faba (KC189947), Vigna radiata (NC_015121) and Vigna angularis (Willd.) Ohwi & H.Ohashi (NC_021092). Syntenic blocks shared by C. australe and other species were identified using both Mauve (ver. 2.3.1, see http://darlinglab.org/mauve/mauve.html; Darling et al. 2004), with default parameters as implemented in Geneious, as well as using BLASTn (ver. 2.8.1, see ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) with a word size of seven and an e-value cutoff of 1 × 10−6, following Guo et al. (2016).
Prediction of RNA editing sites
C-to-U RNA editing sites in the 30 mitochondrial protein coding genes of C. australe and other selected Papilionoideae mitochondrial genomes were predicted using the PREP-Mt online tool (see http://prep.unl.edu/; Mower 2009), with a cutoff value of 0.5, with the exception of six genes (cox2, rpl2, rps1, rps19, sdh3 and sdh4) that were not universally present in Papilionoideae mitochondrial genomes.
Phylogenetic reconstruction of Papilionoideae
To test the utility of mitochondrial genome sequences in resolving the deep relationships of Papilionoideae, phylogenetic analyses were performed using 33 protein-coding genes (atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFc, ccmFn, cob, cox1, cox2, cox3, matR, mttB, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl5, rpl16, rps1, rps3, rps4, rps10, rps12, rps14 and sdh4) of the 10 complete mitochondrial genomes of Papilionoideae, with the mitochondrial genome of Senna tora (L.) Roxb. (Caesalpinioideae; NC_038053) as the outgroup. Genes were aligned using MAFFT (ver. 7.1.2, see https://mafft.cbrc.jp/alignment/software/; Katoh and Standley 2013) using the L-INS-I algorithm. Gene alignments were subsequently adjusted manually and concatenated in Geneious, which produced a 35 360-bp nucleotide dataset. Phylogenetic analyses were performed with standard Bayesian inference (BI) and Maximum likelihood (ML) methods. The best-fit substitution model was estimated in PartitionFinder2 (ver. 2.1.1, see http://www.robertlanfear.com/partitionfinder/; Lanfear et al. 2017), by using the following settings: the all model, RAxML (ver. 8.2.12, see https://github.com/stamatak/standard-RAxML; Stamatakis 2014), the rcluster algorithm (Lanfear et al. 2014) with the rcluster-percent set to 10, and the Akaike information criterion (AICc) to compare the fit of the different models. The BI analysis was performed with MrBayes (ver. 3.2, see http://nbisweden.github.io/MrBayes/download.html; Ronquist et al. 2012), with four Markov Chain Monte Carlo (MCMC) runs using a random starting tree, an invgamma rate model with six discrete categories and 10 million generations, with a sampling frequency of 1 every 1000 generations, and the first 25% being discarded as burn-in. Stationarity was considered to be reached when the average standard deviation of split frequencies was <0.01. Maximum likelihood analysis was performed with RAxML. Branch support was estimated using 1000 rapid bootstrap replicates (‘-f a’ option) with the GTRGAMMA substitution model (Stamatakis 2006).
Results and discussion
Mitochondrial genome assembly and verification
Repeats are likely to mediate intramolecular recombination within mitochondrial genomes, leading to their often-complex genome structure (Palmer and Shields 1984; Alverson et al. 2011b; Cole et al. 2018; Kovar et al. 2018). Such complexity often precludes confident mitochondrial genome assemblage using short inserts and reads, leading to the availability of few angiosperm mitochondrial genomes, including in Papilionoideae. For example, the large repeats of Glycine max are responsible for its complex mitochondrial genome structure (Chang et al. 2013). In sequencing the C. australe mitochondrial genome, we employed paired-end sequencing with a 350-bp insert size to try to maximise our ability to fully assemble the genome by using short (150 bp) reads. Only three repeats >100 bp in length were identified in C. australe, namely, R1 (119 bp), R2 (133 bp) and R3 (114 bp) (Fig. 1). Homologous recombination among these repeats yielded nine hypothetical alternative mitochondrial-genome arrangements (C1−C9; Fig. 1) on the basis of the original assembly graph (OC). Through mapping of paired-end reads, subgenomic conformations C1−C3 and C5−C8 were rejected (Fig. 1) because paired-end reads did not completely span these repeats. In contrast, two circular conformations (C4 and C9) that contained two copies of all three repeats and contained exactly the same genomic information were strongly supported. Conformations C4 and C9 differed only in the orientation of the 64-kb fragment D (Fig. 1). Read mapping against these two confirmations yielded 367 consistent paired-end reads that spanned Repeat R3 (Fig. 1) at the inversion breakpoint in the major conformation (C4, or MC1; Fig. 1), whereas only six paired-end reads consistently covered R3 at the inversion breakpoint in the minor conformation (C9, or MC2; Fig. 1). MC1 and MC2 are depicted as circular chromosomes with lengths of 542 079 bp. A circular map of MC1 is shown in Fig. 2; the sequence of MC1 was deposited in GenBank as accession MK426679. A linear map of both MC1 and MC2 is shown in Fig. 3. Repeats R1, R2 and R3 separate the MC1 and MC2 into six fragments, labelled A (303 789 bp), B (72 549 bp), C (2518 bp), D (64 174 bp), E (64 831 bp) and F (33 486 bp) (Fig. 1). The endpoints of fragment D, which is inverted between MC1 and MC2, are located between trnfM and Exon 1 of nad1 (Fig. 3).
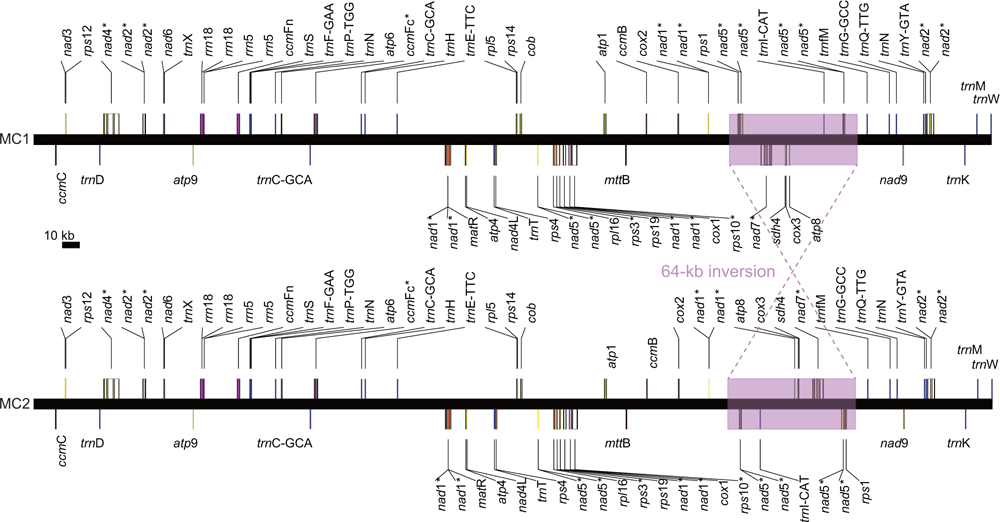
|
Mitochondrial genome features of C. australe
Of the 8 610 483 paired-end reads, 704 104 mapped to the MC1 mitochondrial genome. The mean mitochondrial genome coverage was ~389.7× (Fig. S1, available as Supplementary material to this paper; coverage ranged from ~12× to ~911×). The GC content was 45.3%, which is typical for other selected mitochondrial genomes (Table S1, available as Supplementary material to this paper). The mitochondrial genome of C. australe is large among published Papilionoideae mitochondrial genomes, which a range from 217 618 bp in Medicago truncatula to 588 000 bp in Vicia faba (Table S1). The large size of the mitochondrial genome of C. australe is mainly due to the accumulation of species-specific non-coding sequences, which account for 87.7% (475 542 bp) of the mitochondrial genome. The length of the intergenic regions also contributes greatly to the variation in the mitochondrial genome size among other Papilionoideae species.
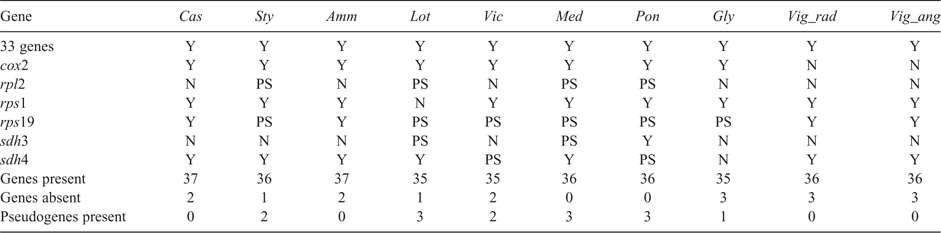
The mitochondrial genome of C. australe comprises 37 known protein-coding genes, three rRNAs and 18 tRNAs (Table 1). Of these, trnC–GCA and trnN are present in two copies. Of the protein-coding genes, 33 are also found in all other available Papilionoideae mitochondrial genomes (Table 1). The cox2 gene is absent from Vigna radiata and V. angularis, and rps1 is absent from Lotus japonicus. The full rRNA and tRNA gene set in C. australe is also shared with all other Papilionoideae species. The total length of the mitochondrial protein-coding gene sequences in C. australe is 30 135 bp, which is comparable to that in most other papilionoid species, but is 11 kb shorter than those of Ammopiptanthus mongolicus and Vicia faba. This is because the mitochondrial genome of A. mongolicus has seven chloroplast-derived protein-coding genes (atpA, atpB, ndhJ, ndhK, psbA, rbcL and rpoB), and the mitochondrial genome of V. faba possesses two or three copies of 10 mitochondrial genes (atp1, atp6, atp9, ccmC, ccmFc, mttB, nad7, nad9, rpl16 and rps3).
Intronic regions comprise 5.4% of the C. australe mitochondrial genome, including nine cis-spliced introns and eight trans-spliced introns (Table S1) across eight protein-coding genes (ccmFc, nad1, nad2, nad4, nad5, nad7, rps3 and rps10). These introns are homologous to those in other Papilionoideae mitochondrial genomes. Variation in total intron length among Papilionoideae (ranging from 27 066 bp in Lotus japonicus to 32 553 bp in Glycine max) was not high compared with variation in mitochondrial genome length. Five introns (ccmFc intron, nad2 Intron 2, nad4 Intron 3, rps10 intron and rps3 intron) showed high intron-length variations across Papilionoideae (Table S1, Fig. S2, available as Supplementary material to this paper); introns within the ccmFc gene of C. australe, Styphnolobium japonicum, Ammopiptanthus mongolicus, L. japonicus, Medicago truncatula and Vicia faba are only ~950 bp, but they are ~4 kb in other sequenced mitochondrial genomes of Papilionoideae. The intron of the rps10 gene also shows high length variation in Papilionoideae; the intron is 1.9 kb shorter in the Hologalegina clade (L. japonicus, Vicia faba and M. truncatula) than in remaining Papilionoideae. The length variations of ccmFc intron and rps10 intron are similar to those found in studies of Chang et al. (2013) and Shi et al. (2018).
RNA editing of Papilionoideae mitochondrial genomes
Plant mitochondrial genomes have a high incidence of C-to-U RNA editing (Hiesel et al. 1989; Takenaka et al. 2008; Alverson et al. 2010; Suzuki et al. 2013), with ~200−800 editing sites in the protein-coding mitochondrial genes of most angiosperms (Mower 2008; Sloan et al. 2010; Richardson et al. 2013; Guo et al. 2016). However, the incidence of RNA editing across lineages of Papilionoideae is largely unknown (Kovar et al. 2018; Shi et al. 2018). Within C. australe, 487 C to U RNA editing sites were predicted among the 30 protein-coding genes (Table S2, available as Supplementary material to this paper). Among these RNA editing sites, 34.5% (168 sites) occurred in the first base position of the codon, 65.5% (319 sites) occurred in the second base position, and none occurred in the third position. Among these protein-coding genes, nad4 possessed the highest number of RNA editing sites (42 sites) and atp1 possessed the fewest (1 sites). In total, 52.8% (257 sites) of the RNA editing sites involved amino acids that were converted from hydrophilic to hydrophobic, whereas 7.6% (37 sites) involved conversion from hydrophobic to hydrophilic amino acids. In contrast, 30.6% (149 sites) involved hydrophobic to hydrophobic amino acid conversions and 8.2% (40 sites) involved hydrophilic to hydrophilic amino acid conversions. Two glycines and two arginines were converted into stop codons. The ccmB, nad4L and ccmC genes had the highest editing frequency, with 5.31, 4.29 and 4.05 edits per 100 nt respectively, whereas the editing frequencies of other genes were lower than four editing sites per 100 nt (Table S3, available as Supplementary material to this paper).
Papilionoideae mitochondrial genes showed RNA editing patterns that were largely similar to those of other angiosperms (Alverson et al. 2010). The positions and amino acid conversions of predicted RNA editing of the 10 mitochondrial genomes in Papilionoideae were provided in Tables S4–S13 (available as Supplementary material to this paper). Across 30 genes that were present in all 10 Papilionoideae mitochondrial genomes, the lowest levels of RNA editing were predicted in atp1 and atp9, with only one or two editing sites. The ndh4 gene had the highest level of editing, with an average of 40.8 editing sites. Variation in the RNA editing-site number across Papilionoideae was highest in atp6, where it ranged from two to 20, whereas four genes (atp4, atp8, rps14, rps4) had the same number of editing sites across all Papilionoideae mitochondrial genomes (Fig. 4, Tables S2, S–S13). Papilionoid ribosomal protein-coding genes had fewer edits, whereas ccmB, nad4L and ccmC had the highest editing frequency.
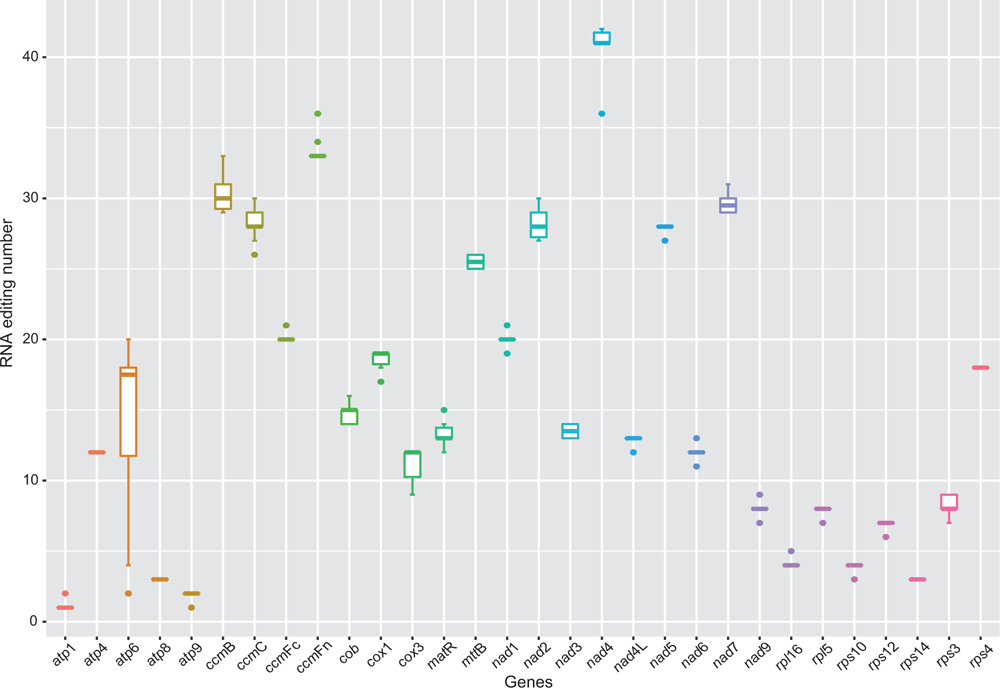
|
Previous studies have shown that closely related taxa generally share more RNA-editing sites (Chen et al. 2011), and we detected a similar phenomenon in Papilionoideae; of the respective 487 and 489 RNA editing sites that were found in the C. australe and Styphnolobium japonicum mitochondrial genes, 473 sites were shared. In Vigna radiata and V. angularis, all 475 detected RNA editing sites were shared. In the four species of the Millettioid clade (Fig. 4), 455 sites were shared between Pongamia pinnata and Glycine max, 456 sites were shared between G. max and Vigna radiata or V. angularis, and 445 sites were shared between P. pinnata and V. radiata or V. angularis.
Phylogenetic analyses of Papilionoideae mitochondrial genomes
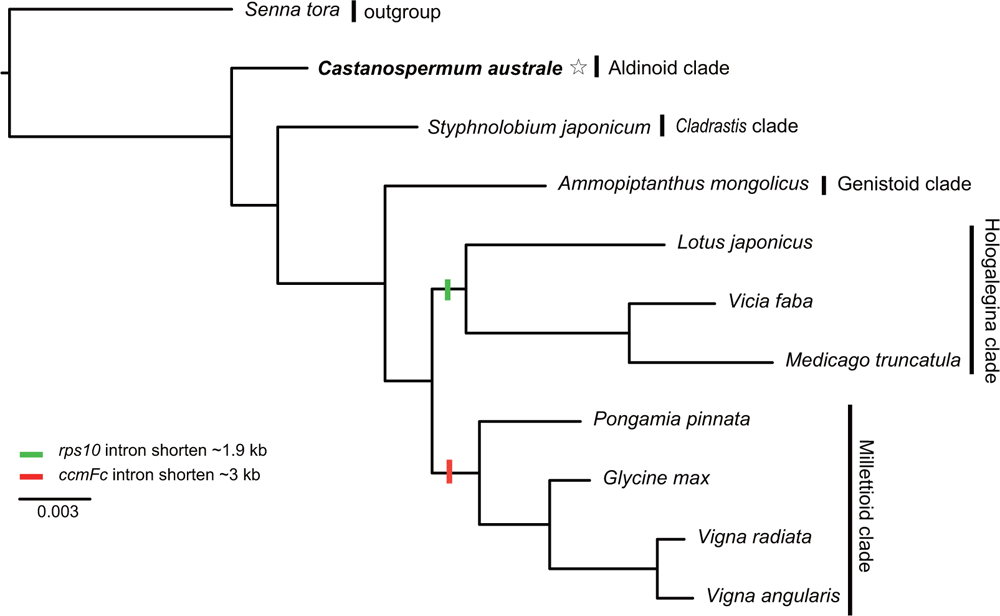
Analyses based on 33 mitochondrial genes resolved the deep phylogenetic relationships of Papilionoideae, with maximum bootstrap support and posterior probability at all nodes (Fig. 5). The Aldinoid clade and Cladrastis clade were successively sister to remaining Papilionoideae. These early diverging Papilionoideae relationships were not resolved in Legume Phylogeny Working Group (2017) on the basis of plastid matK sequence, although recent phylogenomic analyses using 81 plastid-coding genes recovered the same relationships (R. Zhang, Y. H. Wang, J. J. Jin, A. Bruneau, D. Cardoso, L. P. de Queiroz, M. Moore, S. D. Zhang, S. Y. Chen, J. Wang, D. Z. Li, T. S. Yi, unpubl. data). Hence, concatenation of the mitochondrial genes does in fact provide useful information for resolving deep phylogenetic relationships of Papilionoideae.
Synteny
Angiosperm mitochondrial genomes usually have low structural conservation (Knoop 2012; Mower et al. 2012b; Guo et al. 2016). Synteny analyses of Papilionoideae mitochondrial genomes showed extreme structural variations, including a large amount of re-arrangement and inversions (Fig. S3, available as Supplementary material to this paper). Blocks of ~136−261 kb in the mitochondrial genome of C. australe were homologous to those in the remaining nine genomes compared here. However, closely related taxa shared more syntenic blocks. For example, 48.3% (261 763 bp) of the 542 079-bp mitochondrial genome of C. australe was homologous to the more closely related Styphnolobium japonicum, but only 33.5% (181 693 bp) and 33.1% (179 660 bp) were syntenic to the more distantly related Ammopiptanthus mongolicus and Glycine max respectively (Fig. 6). This variation is not surprising, given that recombination events in plant mitochondria are known to play a major role in the structural variation of mitochondrial genomes (Arrieta-Montiel and Mackenzie 2011; Knoop et al. 2011; Kühn and Gualberto 2012), including within Papilionoideae mitochondrial genomes (Chang et al. 2013; Shi et al. 2018).
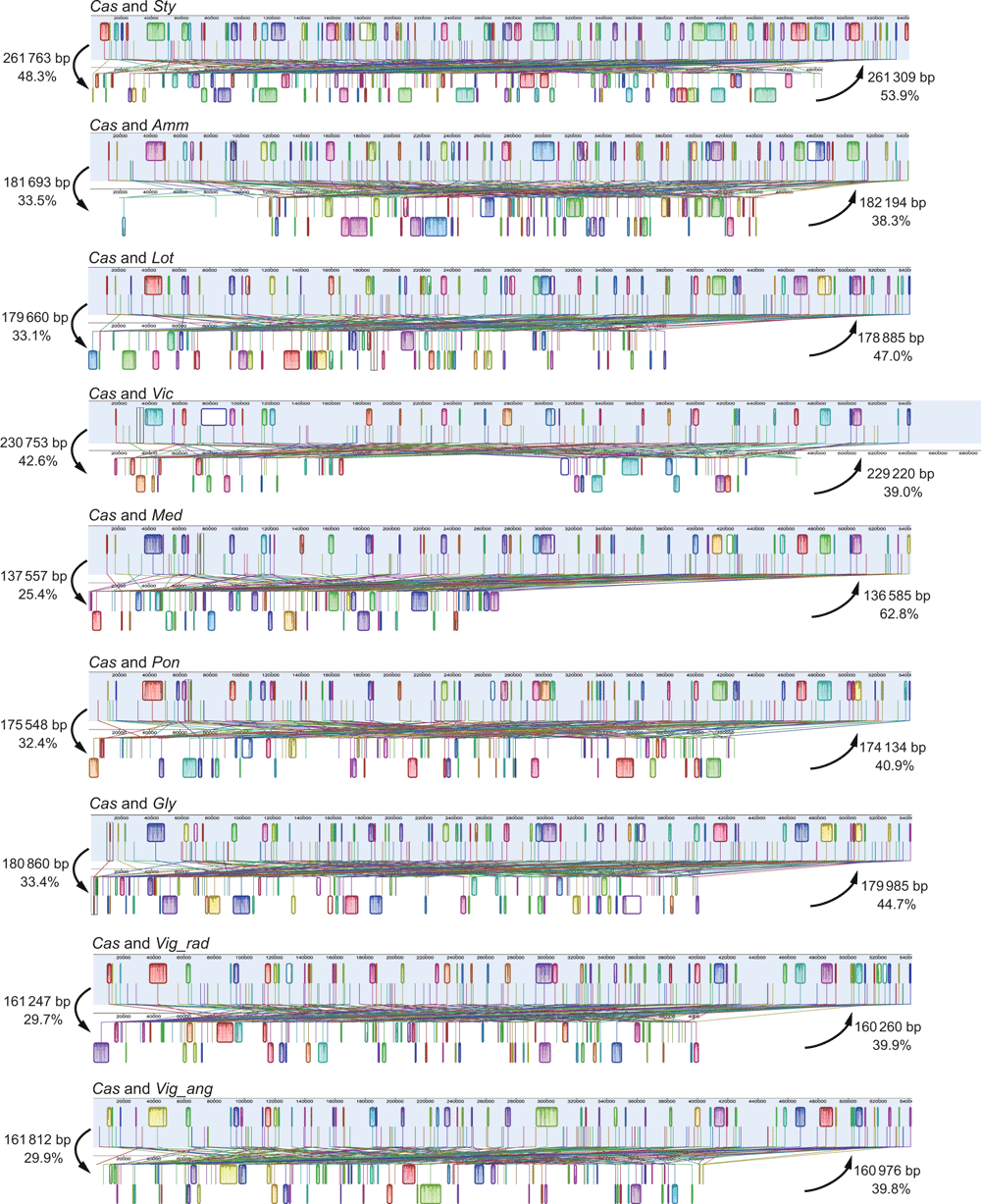
|
Conclusions
The comparison of the C. australe mitochondrial genome with nine other mitochondrial genomes of subfamily Papilionoideae has provided a much-improved understanding of mitochondrial genomic evolution across papilionoid legumes. Although these genomes share strong conservation of gene and intron content as well as some syntenic blocks and similarities in RNA editing, their variable genome size and structure indicate that recombination within Papilionoideae mitochondrial genomes has, nevertheless, been extensive. Equally important, our study demonstrated the value of mitochondrial genes for resolving deep-level phylogenetic relationships in legumes. Future research should continue to target legume mitochondrial genomes for sequencing and assembly, to further develop our understanding of the evolution of this overlooked genome, and to mine it for useful phylogenetic characters.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This work was supported by grants from the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31010000), the National Natural Science Foundation of China [key international (regional) cooperative research project number 31720103903], and the Large-scale Scientific Facilities of the Chinese Academy of Sciences (number 2017-LSF-GBOWS-02).
Acknowledgements
We gratefully acknowledge the Brisbane Botanic Garden, Queensland, Australia, for permission to collect Castanospermum australe, and the Plant Germplasm and Genomics Center, Kunming Institute of Botany, Chinese Academy of Sciences (KIB, CAS), for DNA extraction. We are also grateful to reviewers and the editor for improving the manuscript.
References
Adams KL, Qiu YL, Stoutemyer M, Palmer JD (2002) Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proceedings of the National Academy of Sciences of the United States of America 99, 9905–9912.| Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution.Crossref | GoogleScholarGoogle Scholar | 12119382PubMed |
Alverson AJ, Wei XX, Rice DW, Stern DB, Barry K, Palmer JD (2010) Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Molecular Biology and Evolution 27, 1436–1448.
| Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae).Crossref | GoogleScholarGoogle Scholar | 20118192PubMed |
Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD (2011a) The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One 6, e16404
| The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats.Crossref | GoogleScholarGoogle Scholar | 21283772PubMed |
Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD (2011b) Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. The Plant Cell 23, 2499–2513.
| Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber.Crossref | GoogleScholarGoogle Scholar | 21742987PubMed |
Arrieta-Montiel MP, Mackenzie SA (2011) Plant mitochondrial genomes and recombination. In ‘Plant Mitochondria’. (Ed. F Kempken) pp. 65–82. (Springer Science + Business Media: New York, NY, USA)
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin A, Sirotkin A, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19, 455–477.
| SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.Crossref | GoogleScholarGoogle Scholar | 22506599PubMed |
Bi C, Paterson AH, Wang X, Xu Y, Wu D, Qu Y, Jiang A, Ye Q, Ye N (2016) Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Research International 2016, 5040598
| Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches.Crossref | GoogleScholarGoogle Scholar | 27847816PubMed |
Chang S, Wang Y, Lu J, Gai J, Li J, Chu P, Guan R, Zhao T (2013) The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS One 8, e56502
| The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels.Crossref | GoogleScholarGoogle Scholar | 24367547PubMed |
Chen JM, Guan RZ, Chang SX, Du TQ, Zhang HS, Xing H (2011) Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS One 6, e17662
| Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L.Crossref | GoogleScholarGoogle Scholar | 22096498PubMed |
Cole LW, Guo WH, Mower JP, Palmer JD (2018) High and variable rates of repeat-mediated mitochondrial genome rearrangement in a genus of plants. Molecular Biology and Evolution 35, 2773–2785.
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Research 14, 1394–1403.
| Mauve: multiple alignment of conserved genomic sequence with rearrangements.Crossref | GoogleScholarGoogle Scholar |
Gualberto JM, Newton KJ (2017) Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annual Review of Plant Biology 68, 225–252.
| Plant mitochondrial genomes: dynamics and mechanisms of mutation.Crossref | GoogleScholarGoogle Scholar | 28226235PubMed |
Guo WH, Grewe F, Fan W, Young GJ, Knoop V, Palmer JD, Mower JP (2016) Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Molecular Biology and Evolution 33, 1448–1460.
| Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution.Crossref | GoogleScholarGoogle Scholar |
Guo WH, Zhu AD, Fan WS, Mower JP (2017) Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytologist 213, 391–403.
| Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns.Crossref | GoogleScholarGoogle Scholar |
Hiesel R, Wissinger B, Schuster W, Brennicke A (1989) RNA editing in plant mitochondria. Science 246, 1632–1634.
| RNA editing in plant mitochondria.Crossref | GoogleScholarGoogle Scholar | 2480644PubMed |
Hiesel R, Vonhaeseler A, Brennicke A (1994) Plant mitochondrial nucleic-acid sequences as a tool for phylogenetic analysis. Proceedings of the National Academy of Sciences of the United States of America 91, 634–638.
| Plant mitochondrial nucleic-acid sequences as a tool for phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar | 7507251PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780.
| MAFFT multiple sequence alignment software version 7: improvements in performance and usability.Crossref | GoogleScholarGoogle Scholar | 23329690PubMed |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar | 22543367PubMed |
Knoop V (2012) Seed plant mitochondrial genomes: complexity evolving. In ‘Genomics of Chloroplasts and Mitochondria’. (Ed. R Bock, V Knoop) pp. 175–200. (Springer: Dordrecht, Netherlands)
Knoop V, Volkmar U, Hecht J, Grewe F (2011) Mitochondrial genome evolution in the plant lineage. In ‘Plant Mitochondria’. (Ed. F Kempken) pp. 3–29. (Springer Science + Business Media: New York, NY, USA)
Kovar L, Nageswara-Rao M, Ortega-Rodriguez S, Dugas DV, Straub S, Cronn R, Strickler SR, Hughes CE, Hanley KA, Rodriguez DN, Langhorst BW, Dimalanta ET, Bailey CD (2018) Pacbio-based mitochondrial genome assembly of Leucaena trichandra (Leguminosae) and an intrageneric assessment of mitochondrial RNA editing. Genome Biology and Evolution 10, 2501–2517.
| Pacbio-based mitochondrial genome assembly of Leucaena trichandra (Leguminosae) and an intrageneric assessment of mitochondrial RNA editing.Crossref | GoogleScholarGoogle Scholar | 30137422PubMed |
Kühn K, Gualberto JM (2012) Recombination in the stability, repair and evolution of the mitochondrial genome. Mitochondrial Genome Evolution 63, 215–252.
| Recombination in the stability, repair and evolution of the mitochondrial genome.Crossref | GoogleScholarGoogle Scholar |
Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A (2014) Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evolutionary Biology 14, 82–95.
| Selecting optimal partitioning schemes for phylogenomic datasets.Crossref | GoogleScholarGoogle Scholar | 24742000PubMed |
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34, 772–773.
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359.
| Fast gapped-read alignment with Bowtie 2.Crossref | GoogleScholarGoogle Scholar | 22388286PubMed |
Legume Phylogeny Working Group (2013) Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248.
| Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades.Crossref | GoogleScholarGoogle Scholar |
Legume Phylogeny Working Group (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77.
| A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny.Crossref | GoogleScholarGoogle Scholar |
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41, W575–W581.
| OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets.Crossref | GoogleScholarGoogle Scholar | 23609545PubMed |
Maréchal A, Brisson N (2010) Recombination and the maintenance of plant organelle genome stability. New Phytologist 186, 299–317.
| Recombination and the maintenance of plant organelle genome stability.Crossref | GoogleScholarGoogle Scholar | 20180912PubMed |
Mower JP (2008) Modeling sites of RNA editing as a fifth nucleotide state reveals progressive loss of edited sites from angiosperm mitochondria. Molecular Biology and Evolution 25, 52–61.
| Modeling sites of RNA editing as a fifth nucleotide state reveals progressive loss of edited sites from angiosperm mitochondria.Crossref | GoogleScholarGoogle Scholar | 17940211PubMed |
Mower JP (2009) The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Research 37, W253–W259.
| The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments.Crossref | GoogleScholarGoogle Scholar | 19433507PubMed |
Mower JP, Case AL, Floro ER, Willis JH (2012a) Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biology and Evolution 4, 670–686.
| Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS.Crossref | GoogleScholarGoogle Scholar | 22534162PubMed |
Mower JP, Sloan DB, Alverson AJ (2012b) Plant mitochondrial genome diversity: the genomics revolution. In ‘Plant Genome Diversity’. (Ed. JF Wendel) pp. 123–144. (Springer: Vienna, Austria)
Naito K, Kaga A, Tomooka N, Kawase M (2013) De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breeding Science 63, 176–182.
| De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers.Crossref | GoogleScholarGoogle Scholar | 23853512PubMed |
Negruk V (2013) Mitochondrial genome sequence of the legume Vicia faba. Frontiers in Plant Science 4, 128
| Mitochondrial genome sequence of the legume Vicia faba.Crossref | GoogleScholarGoogle Scholar | 23675376PubMed |
Ostrander GK, Scribner NK, Rohrschneider LR (1988) Inhibition of V-Fms-induced tumor-growth in nude-mice by castanospermine. Cancer Research 48, 1091–1094.
Palmer JD, Shields CR (1984) Tripartite structure of the Brassica campestris mitochondrial genome. Nature 307, 437–440.
| Tripartite structure of the Brassica campestris mitochondrial genome.Crossref | GoogleScholarGoogle Scholar |
Palmer JD, Adams KL, Cho YR, Parkinson CL, Qiu YL, Song KM (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proceedings of the National Academy of Sciences of the United States of America 97, 6960–6966.
| Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates.Crossref | GoogleScholarGoogle Scholar | 10860957PubMed |
Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD (2013) The ‘fossilized’ mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11, 29
| The ‘fossilized’ mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate.Crossref | GoogleScholarGoogle Scholar | 23587068PubMed |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
| MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Crossref | GoogleScholarGoogle Scholar | 22357727PubMed |
Shi YC, Liu Y, Zhang SZ, Zou R, Tang JM, Mu WX, Peng Y, Dong SS (2018) Assembly and comparative analysis of the complete mitochondrial genome sequence of Sophora japonica ‘JinhuaiJ2’. PLoS One 13, e0202485
| Assembly and comparative analysis of the complete mitochondrial genome sequence of Sophora japonica ‘JinhuaiJ2’.Crossref | GoogleScholarGoogle Scholar |
Skippington E, Barkman TJ, Rice DW, Palmer JD (2015) Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proceedings of the National Academy of Sciences of the United States of America 112, E3515–E3524.
| Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes.Crossref | GoogleScholarGoogle Scholar | 26100885PubMed |
Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR (2010) Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics 185, 1369–1380.
| Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force.Crossref | GoogleScholarGoogle Scholar | 20479143PubMed |
Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR (2012) Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biology 10, e1001241
| Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates.Crossref | GoogleScholarGoogle Scholar | 22272183PubMed |
Smith DR, Keeling PJ (2015) Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proceedings of the National Academy of Sciences of the United States of America 112, 10177–10184.
| Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes.Crossref | GoogleScholarGoogle Scholar | 25814499PubMed |
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
| RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models.Crossref | GoogleScholarGoogle Scholar | 16928733PubMed |
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
| RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Crossref | GoogleScholarGoogle Scholar | 24451623PubMed |
Sunkara PS, Bowlin TL, Liu PS, Sjoerdsma A (1987) Antiretroviral activity of castanospermine and deoxynojirimycin, specific inhibitors of glycoprotein processing. Biochemical and Biophysical Research Communications 148, 206–210.
| Antiretroviral activity of castanospermine and deoxynojirimycin, specific inhibitors of glycoprotein processing.Crossref | GoogleScholarGoogle Scholar | 2960321PubMed |
Suzuki H, Yu JW, Ness SA, O’Connell MA, Zhang JF (2013) RNA editing events in mitochondrial genes by ultra-deep sequencing methods: a comparison of cytoplasmic male sterile, fertile and restored genotypes in cotton. Molecular Genetics and Genomics 288, 445–457.
| RNA editing events in mitochondrial genes by ultra-deep sequencing methods: a comparison of cytoplasmic male sterile, fertile and restored genotypes in cotton.Crossref | GoogleScholarGoogle Scholar | 23812672PubMed |
Takenaka M, Verbitskly D, van der Merwe JA, Zehrmann A, Brennicke A (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8, 35–46.
| The process of RNA editing in plant mitochondria.Crossref | GoogleScholarGoogle Scholar | 18326075PubMed |
Walker BD, Kowalski M, Goh WC, Kozarsky K, Krieger M, Rosen C, Rohrschneider L, Haseltine WA, Sodroski J (1987) Inhibition of human-immunodeficiency-virus syncytium formation and virus-replication by castanospermine. Proceedings of the National Academy of Sciences of the United States of America 84, 8120–8124.
| Inhibition of human-immunodeficiency-virus syncytium formation and virus-replication by castanospermine.Crossref | GoogleScholarGoogle Scholar | 2825177PubMed |
Wick RR, Schultz MB, Zobel J, Holt KE (2015) Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352.
| Bandage: interactive visualization of de novo genome assemblies.Crossref | GoogleScholarGoogle Scholar | 26099265PubMed |
Yang JB, Li DZ, Li HT (2014) Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Molecular Ecology Resources 14, 1024–1031.
Yu TQ, Sun LC, Cui HW, Liu SL, Men JY, Chen SL, Chen YZ, Lu CF (2018) The complete mitochondrial genome of a tertiary relict evergreen woody plant Ammopiptanthus mongolicus. Mitochondrial DNA – B. Resources 3, 9–11.
| The complete mitochondrial genome of a tertiary relict evergreen woody plant Ammopiptanthus mongolicus.Crossref | GoogleScholarGoogle Scholar |