Molecular and morphological analyses support recognition of Prostanthera volucris (Lamiaceae), a new species from the Central Tablelands of New South Wales
Ryan P. O’Donnell A B E * , Jeremy J. Bruhl
A B E * , Jeremy J. Bruhl  A , Ian R. H. Telford
A , Ian R. H. Telford  A , Trevor C. Wilson
A , Trevor C. Wilson  B , Heidi C. Zimmer
B , Heidi C. Zimmer  C , Guy M. Taseski
C , Guy M. Taseski  D and Rose L. Andrew
D and Rose L. Andrew  A *
A *
A Botany and N.C.W. Beadle Herbarium, School of Environmental and Rural Science, University of New England, Armidale, NSW 2351, Australia.
B National Herbarium of New South Wales, Australian Institute of Botanical Science, Royal Botanic Gardens and Domain Trust, Locked Bag 6002, Mount Annan, NSW 2567, Australia.
C Australian National Herbarium, Centre for Australian National Biodiversity Research, GPO Box 1700, Canberra, ACT 2601, Australia.
D John T. Waterhouse Herbarium, School of Biological, Earth and Environmental Sciences, University of New South Wales, Kensington, NSW 2052, Australia.
E Present address: Division of Ecology and Evolution, Research School of Biology, The Australian National University, Canberra, ACT 2600, Australia.
Australian Systematic Botany 36(1) 1-20 https://doi.org/10.1071/SB22017
Submitted: 20 June 2022 Accepted: 8 January 2023 Published: 8 February 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Research into the systematics of Prostanthera recently revealed close evolutionary relationship among P. phylicifolia sens. str., the critically endangered P. gilesii, and a population of uncertain identity from the Central Tablelands of New South Wales (NSW), Australia. Previous analyses were unable to establish whether genetic boundaries separated these taxa. This study assessed species boundaries among these three taxa by using a combination of single-nucleotide polymorphisms (SNPs) sampled at the population-scale and multivariate analysis of morphological characters. Ordination, model-based clustering, F-statistics, neighbour-network analysis, phylogenetic analysis, and ancestry coefficient estimates all provided support for discrete genetic differences among the three taxa. Morphological phenetic analysis recovered congruent morphological clusters and identified a suite of corresponding diagnostic characters. This congruence of molecular and morphological evidence supports the presence of three independently evolving lineages, two of which correspond with the previously described P. gilesii and P. phylicifolia sens. str. The third taxon, represented by a single population from the Central Tablelands of NSW, is here described as P. volucris R.P.O’Donnell. A detailed description, diagnostic line drawings and photographs are provided. We evaluate P. volucris as satisfying criteria to be considered Critically Endangered.
Keywords: critically endangered, DArTseq, genotyping-by-sequencing, Lamiaceae, population genomics, species delimitation, systematics, taxonomy.
Introduction
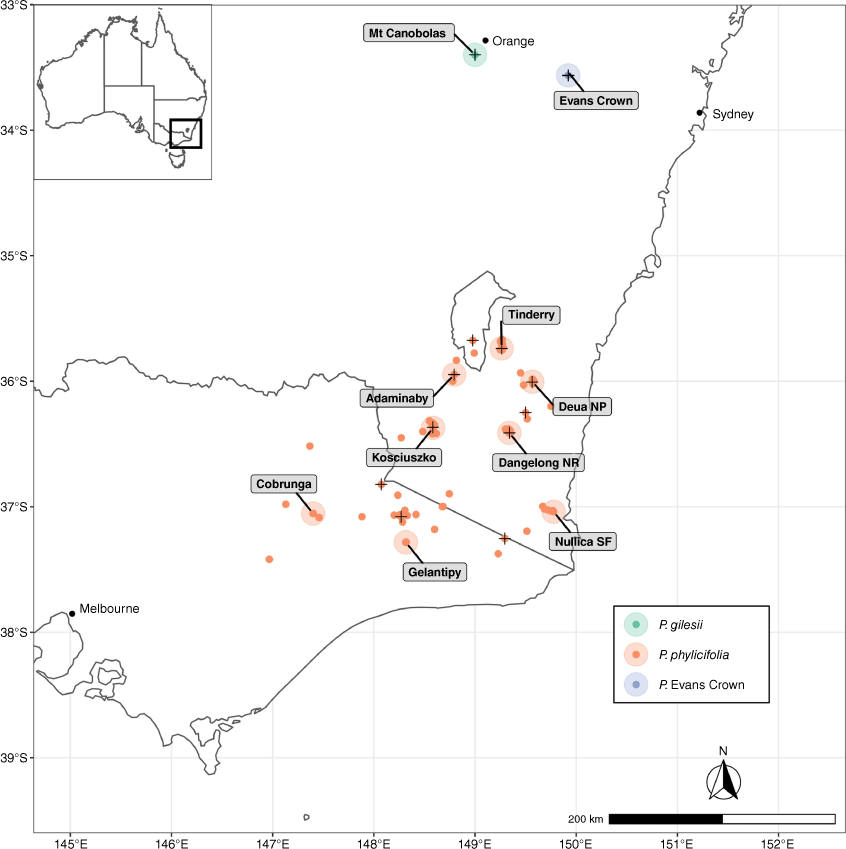
Prostanthera Labill. is the most speciose genus of endemic Australian Lamiaceae, encompassing over 105 accepted species (Conn 1984, 1988; Australian Plant Census 2021; Conn et al. 2021). Recent studies have shown the genus to be far more diverse than currently recognised, with several species complexes that require resolution (Wilson et al. 2012, 2019; Conn et al. 2016, 2021; O’Donnell et al. 2021a). Many species of Prostanthera are niche specialists and tend to grow on isolated, rocky outcrops (Conn 1984, 1988; Conn and Wilson 2015; Wilson and Conn 2015; Wilson et al. 2019). Like other rocky outcrop specialists, these species have highly restricted ranges and are particularly vulnerable to environmental threats (Fitzsimons and Michael 2017; Selwood and Zimmer 2020; Hopper et al. 2021; Silveira et al. 2021). Among the accepted species, 19 are listed as Threatened on the Environment Protection and Biodiversity Conservation Act List of Threatened Flora, representing almost one-fifth of the genus (see https://www.environment.gov.au/cgi-bin/sprat/public/publicthreatenedlist.pl?wanted=flora). One example is the Critically Endangered P. gilesii G.W.Althofer ex B.J.Conn & T.C.Wilson, which is geographically restricted in comparison with its close relative, P. phylicifolia F.Muell. Prostanthera gilesii is currently the subject of a targeted conservation and management project (NSW Department of Planning and Environment 2019; Scott and Auld 2020; H. C. Zimmer, J. J. Bruhl and R. L. Andrew, unpubl. data). The species is currently known only from two small subpopulations within the Mount Canobolas State Conservation Area, south-west of Orange in the Central Tablelands of New South Wales (Conn and Wilson 2015).
Molecular phylogenies recovered a close relationship between P. gilesii and P. phylicifolia s. str., whose distribution extends across the Victorian Alps and Snowy Mountains, Monaro, and Southern Tablelands of New South Wales (O’Donnell et al. 2021a) (Fig. 1). Prostanthera phylicifolia was originally described by von Mueller (1858) from material collected near Omeo in northern Victoria; however, the name has been misapplied since Bentham’s (1870) circumscription of the species, which cited additional specimens from the New England region of New South Wales, and the Glass House Mountains, Queensland. Although specimens variously identified as P. phylicifolia may share glabrous, narrow ovate leaves, populations from Victoria and the Southern Tablelands of New South Wales differ substantially in their floral morphology from populations from the Northern Tablelands of New South Wales and southern Queensland. Southern populations have white corollas with purple and yellow punctate markings that are conspicuously zygomorphic, and anthers with a distinctly elongated connective tissue appendage between both pollen sacks. Conversely, northern populations exhibit entirely mauve corollas that are less strongly zygomorphic, and anthers with appendages that are reduced to absent. Wilson et al. (2017) demonstrated that these distinct floral types were correlated with different pollinator assemblages, and, moreover, indicative of phylogenetic relationship. Thus, species exhibiting differences in floral type were unlikely to be closely related. These results were supported by molecular phylogenies presented by O’Donnell et al. (2021a), which unequivocally demonstrated that P. phylicifolia sens. str. is restricted to Victoria and the Southern Tablelands of New South Wales, whereas populations from the Northern Tablelands of New South Wales and southern Queensland variously identified as P. phylicifolia represent an undescribed taxon more closely allied to P. scutellarioides (R.Br.) Briq. All further references to P. phylicifolia in this manuscript will refer to P. phylicifolia sens. str. as outlined above.
|
Another population of uncertain identification from Evans Crown Nature Reserve, located ~2.8 km south-east of Tarana, New South Wales, was also included in the study and recovered as sister to P. gilesii. Although superficially similar to P. phylicifolia and P. gilesii, the Evans Crown population is distinguishable in a number of ways that suggest it may also be a distinct species. The Evans Crown population differs substantially with respect to several reproductive and vegetative characters (O’Donnell et al. 2021a), and is found on granite outcrops (NSW National Parks and Wildlife Service 2009; R. P. O’Donnell, pers. obs., 2020), whereas its closest relative (P. gilesii) is found on basaltic substrates (Scott and Auld 2020). The differences exhibited by the Evans Crown population do not match diagnoses of P. phylicifolia or P. gilesii (von Mueller 1858; Conn and Wilson 2015). Tree topologies recovered by O’Donnell et al. (2021a) based on cpDNA or nDNA were not sufficiently resolved to confidently assess whether P. phylicifolia was reciprocally monophyletic or paraphyletic with respect to the P. gilesii-Evans Crown clade. Consequently, these data were considered insufficient to determine species-level boundaries.
The aim of this study was to elucidate the relationship among P. gilesii, P. phylicifolia and the Evans Crown population and to assess whether they each represent independently evolving lineages that warrant species-level recognition. Multiple individuals per population were sampled for each putative taxon to produce single-nucleotide polymorphism (SNP) data, which were then analysed using population genetic and phylogenetic methods. Morphometric phenetic analyses were then applied to objectively assess interspecific morphological variation and distinguish informative characters for identification.
Materials and methods
Integrative taxonomic approach
Integrative taxonomy aims to incorporate and synthesise multiple, independent lines of evidence to rigorously test and corroborate delimitation hypotheses, thereby increasing the probability that a set of independently evolving metapopulation lineages, i.e. species, sensu de Queiroz (2007), will be resolved (Dayrat 2005; Padial et al. 2010; Schlick-Steiner et al. 2010). To recover a stable taxonomic classification that agrees with evolutionary history, and to avoid the possibility of taxonomic over-inflation (Georges et al. 2018; Hundsdoerfer et al. 2019), an integrative taxonomic approach must be applied to questions of species delimitation in Prostanthera. Schlick-Steiner et al. (2010) provided a framework for an integrative taxonomic approach, suggesting that morphology be used as a primary line of evidence, followed by a genetic discipline. Previous studies of Prostanthera have incorporated phenetic analyses of morphological characters to rigorously assess phenotypic variation among putative taxa (Conn 1984; Conn et al. 2013, 2021; Wilson et al. 2017). Previous molecular studies of Prostanthera incorporating Sanger sequencing data have provided some species-level resolution (Wilson et al. 2012; Conn et al. 2013, 2016, 2021); however, O’Donnell et al. (2021a) and Wilson et al. (2012) recovered discordant topologies between nuclear and chloroplast datasets, suggesting that hybridisation, introgression, or incomplete lineage sorting may have occurred within Prostanthera. The use of high-throughput sequencing molecular approaches capable of detecting genome-wide admixture was recommended as a means for future studies of Prostanthera to mitigate the confounding effect of these evolutionary processes (O’Donnell et al. 2021a).
Genotyping-by-sequencing
DArTseq (Kilian et al. 2012) is a cost-competitive genotyping-by-sequencing (GBS) platform (Diversity Arrays Technology Pty Ltd (DArT), Canberra, ACT, Australia) that captures SNP data that have provided resolution and demographic inference at the population scale in taxonomically recalcitrant plant groups (Sansaloni et al. 2010; Steane et al. 2011; Joyce et al. 2021; Collins et al. 2022; Wilson et al. 2022). Genotyping-by-sequencing allows for a targeted fraction of an organism’s genome to be sequenced in species with little pre-existing reference information (Narum et al. 2013; Soltis et al. 2013; Fernández-Mazuecos et al. 2017). In contrast to microsatellite or amplified fragment-length polymorphism (AFLP) approaches, which rely only on a small number of neutral molecular markers representing a limited subset of the genome, GBS approaches greatly increase the number of putatively neutral markers assayed, thereby improving the precision of estimates of population structure, admixture, and demography (Narum et al. 2013). DNA extraction, library preparation and genotyping-by-sequencing using the DArTseq platform were conducted by DArT. The resulting reads were filtered, assembled de novo and scored by DArT internally, using their proprietary analysis pipeline following methods similar to those outlined by Kilian et al. (2012) and Cruz et al. (2013).
Sampling
Material was sourced from herbarium specimens or field collections of silica-dried or freeze-dried leaf material. Prostanthera phylicifolia was collected from 12 populations spanning its geographic distribution, P. gilesii was collected from both known subpopulations within the Mount Canobolas State Conservation Area (here referred to as Towac and Walls), and material was collected from the single known population located within the Evans Crown Nature Reserve (Fig. 1). Leaf material was also sampled from a herbarium voucher from Evans Crown (Rodd 11009, NSW 856887), collected from coordinates that do not match the immediate vicinity of the known population; however, the DNA yield for this specimen was too low for sequencing. The compact and tangled habit of P. gilesii and the Evans Crown taxon complicated an accurate estimation of the number of individuals. Samples were collected from plants at least 6 m apart to reduce repeated sampling of the same genet, but where separate individuals were clear, each was sampled (e.g. the Walls population consists of two individuals within a shared 3-m-diameter space). Herbarium vouchers were lodged at the National Herbarium of New South Wales (NSW) and the N.C.W. Beadle Herbarium (NE). Flowering material was collected and preserved in 70% ethanol for morphological examination.
To recover SNP data for population genetic analyses, 125 samples were sequenced and co-analysed by DArT. Of the 125 samples, 30 samples from the Evans Crown population, 7 samples of P. phylicifolia and 1 sample of P. gilesii (Walls) were newly sequenced for this study. New samples were co-analysed with samples of P. gilesii (62), P. phylicifolia (13), and the Evans Crown population (7) that had been previously sequenced as part of targeted conservation and management projects underway for P. gilesii (NSW Department of Planning and Environment 2019; Zimmer et al., unpubl. data). Three samples from the same preliminary P. gilesii sequencing round were sequenced again as technical duplicates (Supplementary Table S1). One sample of P. phylicifolia that had previously been sampled as part of a targeted conservation and management project for P. densa A.A.Ham. and P. marifolia R.Br. was included in this co-analysis (Yap et al. 2020).
To provide outgroup representatives for phylogenetic analysis, an additional SNP dataset was co-analysed by DArT where selected samples of P. gilesii (4), P. phylicifolia (14) and the Evans Crown population (3) were co-analysed with samples of P. densa (2), P. marifolia (3), P. granitica Maiden & Betche (2) and P. scutellarioides (R.Br.) Briq. (2) that had previously been sampled as part of a targeted conservation and management project for P. densa and P. marifolia (Yap et al. 2020). These species were chosen as outgroup representatives because they were previously recovered within clades that were sister to the clade containing P. gilesii, P. phylicifolia and the Evans Crown population (O’Donnell et al. 2021a). In total, 30 samples were included for this analysis.
For morphometric phenetic analysis, 20 herbarium voucher specimens were measured and scored (Fig. 1, Supplementary Table S2). For both P. gilesii and the Evans Crown population, five voucher specimens were scored. For P. phylicifolia, 10 specimens were scored to cover the species’ broader distribution. Each herbarium voucher specimen was considered as an individual plant and OTU when scoring. Voucher specimens were selected to incorporate the extent of variability within a taxon, and on the basis that they could provide three replicates for each character being scored.
Fieldwork was conducted under New South Wales Department of Planning and Environment Scientific Licence SL100305.
Molecular analysis of SNP data
The SNP dataset delivered by DArT was processed with the package dartR (ver. 2.0.4, see https://cran.r-project.org/package=dartR; Gruber et al. 2018; Mijangos et al. 2022) in the statistical package R (ver. 4.2.0, R Foundation for Statistical Computing: Vienna, Austria, see https://www.r-project.org/). Processing followed methods outlined by Gruber et al. (2018). First, the ‘gl.filter.secondaries’ function was used to remove secondary SNPs (i.e. sequenced fragments with more than one SNP). This function keeps one SNP from each fragment identified as having multiple SNPs. The SNP dataset was then filtered on the basis of read depth using the ‘gl.filter.rdepth’ function with the default settings. Sites were then filtered to require 98% reproducibility (as estimated by the DArT proprietary pipeline) by using ‘gl.filter.reproducibility’. The remaining loci were filtered on the basis of a minimum call rate of 95% by using gl.filter.callrate. Individuals were then filtered on the basis of individual call rate with the same function, requiring a minimum call rate of 80%. The resultant dataset comprised 10 486 loci scored for 120 individuals, i.e. 5 individuals were removed by filtering (Table S1). Filtering methods were the same for the secondary dataset used for phylogenetic analysis, with the exception of filters for loci and individual call rates. The minimum locus call rate was lowered to 80% to keep as many loci as possible and the minimum individual call rate was lowered to 60% to ensure that outgroup representatives were not filtered out. A total of 2 individuals were removed by filtering and the resultant dataset for phylogenetic analysis comprised 2519 loci scored for 27 individuals (Table S1).
Exclusion of clones and close relatives helps avoid bias in population genetic parameters in species with mixed sexual and asexual reproduction (Montalvo et al. 1997). To identify putative clones and mitigate their confounding effect in calculations of population statistics, the function ‘gl.propShared’ was used to calculate a similarity matrix for individuals for each population on the basis of their proportion of shared alleles. Pairwise shared allele frequencies between accessions that had a technical replicate sequenced were used to determine a threshold by which identical (i.e. clonal) accessions could be identified. The most conservative frequency of 0.9910912, between JJB3602ee and JJB3602ee.1, was used as the final cut-off threshold. Pairwise shared allele frequencies above the cut-off threshold were then counted for each accession. Accessions that returned zero pairwise shared allele frequencies above the cut-off were retained as distinct genotypes. The remaining non-zero accessions were then sorted into groups (distinct count value categories) by identifying accessions with the same number of total pairwise frequencies above the cut-off threshold.
The number of distinct count value categories was subtracted from the number of individuals sampled for that population, and count categories with counts below this number were each considered to represent genotypes with several clones present. For each remaining count category, the individual with the lowest mean pairwise frequency was retained for further analyses. Using pairwise shared allele frequencies, 58 samples of P. gilesii, 2 samples of P. phylicifolia and 20 samples of the Evans Crown population were excluded (Table S1) from further analyses. A detailed walkthrough of this process and the associated R script is available at https://github.com/rpodonnell/ASB_PEC.
To assess the similarities among populations and individuals, ordination of the dataset using principal-component analysis (PCA) was conducted using the ‘gl.pcoa’ function in dartR. To perform a neighbour-net network (Bryant and Moulton 2004) analysis and visualise a distance-based network, a Euclidian distance matrix was calculated using the ‘gl.dist.ind’ function in dartR and exported using the ‘splitstree’ function in RSplitsTree (ver. 0.1.0, B. Bickel and T. Zakharko, see https://github.com/IVS-UZH/RSplitsTree). The resulting distance matrix file was then imported to SplitsTree4 (ver. 4.19.0, see https://software-ab.cs.uni-tuebingen.de/download/splitstree4/welcome.html; Huson 1998; Huson and Bryant 2006) where a neighbour-net network was calculated using the ordinary least squares (OLS) variance method and a lambda fraction of 1.0.
To produce a concatenated SNP matrix for phylogenetic analysis, SNP data were exported using the ‘gl2svdquartets’ function in dartR by using ‘Method 2’, which outputs a single line per sample and codes heterozygous SNPs or ambiguities by using standard ambiguity codes. To examine phylogenetic relationships, trees were estimated from the concatenated SNP matrix under a coalescent model by using SVDquartets (Chifman and Kubatko 2014, 2015) as implemented in PAUP* (ver. 4.0a169; Swofford 2002). Quartets were sampled exhaustively by using the Quartet FM (QFM) quartet-assembly algorithm, with 1000 standard bootstrap replications conducted to estimate branch support. Bootstrap support values were considered strong if they provided support values of ≥95%, moderate from 80 to 94% and weak from 50 to 79%. The final bootstrap consensus tree was visualised using the R package ggtree (ver. 3.6.1, see https://doi.org/doi:10.18129/B9.bioc.ggtree; Yu et al. 2017; Xu et al. 2022). Tree files output by PAUP*, including the final bootstrap consensus tree and all bootstrap replicate trees, are included in the supplementary dataset (available at https://hdl.handle.net/1959.11/53809).
To assess levels of genetic diversity, observed heterozygosity (HO) and expected heterozygosity (HE) values were calculated using the ‘gl.report.heterozygosity’ function in dartR, which calculates SNP heterozygosity. Statistics were calculated at putative species level on the basis of clusters observed in PCA and neighbour-network analyses, by using the unbiased estimate of HE to account for limited and variable sample sizes. Statistics were also calculated for populations with five or more samples. To assess levels of genetic divergence, pairwise FST values were calculated for putative species groups, and between populations with five or more samples using the ‘stamppFst’ function in the R package StAMPP (ver. 1.6.3, see https://cran.r-project.org/package=StAMPP; Pembleton et al. 2013), which estimates pairwise FST values according to Weir and Cockerham (1984). To estimate statistical support, 1000 bootstrap replicates and confidence intervals were calculated. Confidence intervals were estimated using the percentile method, whereby a percentage (default 95%) of bootstrapped FST values around the mean FST are selected, and the minimum and maximum values are recorded as upper and lower confidence intervals (Pembleton et al. 2013). Both subpopulations of P. gilesii were treated as a single population in these calculations. Other populations with five or more samples that were used in calculations included the Dangelong Nature Reserve and Adaminaby populations of P. phylicifolia, and the Evans Crown population. FST values of <0.05 were considered to indicate low genetic differentiation, 0.05–0.25 indicated moderate genetic differentiation, and values >0.25 indicated pronounced differentiation (Freeland et al. 2011).
To investigate admixture, individual ancestry coefficients were estimated using sparse non-negative matrix factorisation (sNMF) in the R package LEA (ver. 3.10.0, see https://bioconductor.org/packages/release/bioc/html/LEA.html; Frichot and François 2015). The package uses cross-entropy values to infer the probable number of ancestral populations (K) in the data, and to assign individuals to genetic clusters. The optimal number of ancestral populations was selected on the basis of the post-stabilisation of the steepest decline in cross-entropy values (Frichot and François 2015; van der Merwe et al. 2021). The ‘snmf’ function was executed for values of K = 1–8, with 50 replicates for each value of K.
Phenetic analysis of morphological data
The character list for morphometric phenetic analysis (Supplementary Table S3) consisted of 29 morphological characters, comprising 12 vegetative and 17 reproductive characters. Characters were selected from previous morphological studies of Prostanthera (Conn 1984; Williams et al. 2006; Conn et al. 2013), with additional characters being added following preliminary examination of specimens (i.e. indumentum direction and density). The mean of replicates (3 per specimen) for quantitative characters (23 in total) was used in morphometric analysis.
Reproductive characters included only calyx, prophyll and podium characters and did not include androecial or gynoecial characters because an insufficient number of specimens exhibited useful reproductive material to score such characters consistently. Measurements of vegetative characters were taken from dry herbarium vouchers. Calyx and prophyll characters were measured from rehydrated specimens except for three specimens of the Evans Crown population (Taseski 853, O’Donnell 30, O’Donnell 55) and one specimen of P. phylicifolia (O’Donnell 61), where calyx and prophyll characters were scored from flowers preserved in 70% ethanol. The final matrix (Supplementary Table S4) contained 29 characters scored for 20 specimens, including 23 quantitative and 6 qualitative characters.
The morphological data matrix was analysed in PATN (ver. 4.0, see https://patn.org/; Belbin and Collins 2013) by using default settings. All characters were weighted equally in each analysis with the Gower association metric, because it is suited to datasets that contain a combination of quantitative, binary, and qualitative characters (Sokal 1986). The unweighted pair-group method using arithmetic means (UPGMA) was used for cluster analysis because it is considered to accurately represent real distances between individuals in taxonomic datasets (Chappill and Ladiges 1992; Copeland et al. 2007). Semi-strong hybrid multidimensional scaling (SSH-MDS) was used for ordination analyses because it has been shown to accurately reflect phenetic patterns (Minchin 1987) and has frequently been used in recent morphological phenetic studies (Plunkett et al. 2009; Bean 2014; de Salas and Schmidt-Lebuhn 2018), including studies of Prostanthera (Conn et al. 2013, 2021).
Results
Principal-component analysis of SNP data
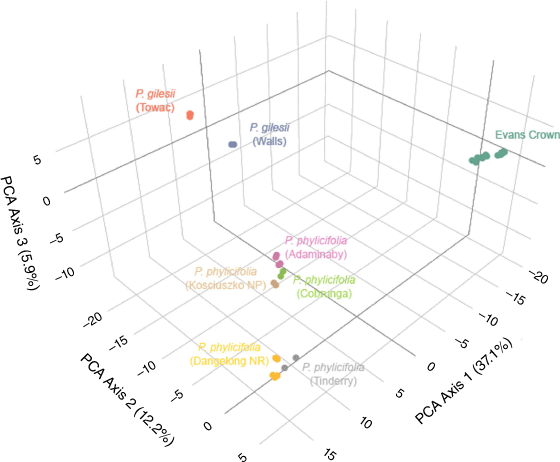
PCA ordination of SNP data (Fig. 2) organised samples as the following three general clusters: (1) all samples of the Evans Crown population as a single cluster; (2) all samples of P. gilesii, subdivided into two subclusters corresponding to their respective subpopulation of origin; and (3) all sampled populations of P. phylicifolia, subdivided into subclusters corresponding to their respective population of origin. Of the first three axes, PCA Axis 1 explained 37.1% of variation, PCA Axis 2 explained 12.2% and PCA Axis 3 explained 5.9%.
|
Neighbour-net network analysis
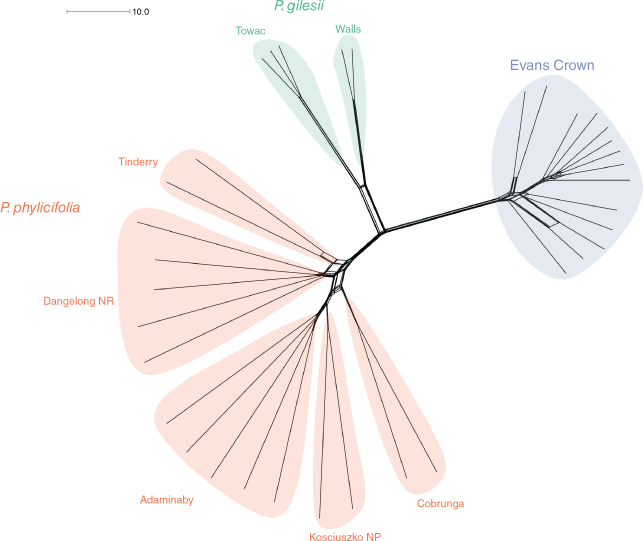
The neighbour-net network graph (Fig. 3) showed three main branches exhibiting little to no reticulation between them. One branch included all samples of the Evans Crown population with some reticulation present among individuals. Another branch included all samples of P. phylicifolia and exhibited limited reticulation among individuals, but some reticulation between populations. Reticulation was observed within two main groups, namely, one consisting of individuals from Tinderry and Dangelong Nature Reserve, and another consisting of individuals from Adaminaby, Kosciuszko National Park and Cobrunga. The third branch included all individuals of P. gilesii, with both specimens from the Walls population forming a distinct subbranch. Little reticulation was present among individuals or between the two subbranches of P. gilesii.
|
Phylogenetic analysis
Exhaustive quartet sampling sampled 17 550 quartets in total, with a compatible quartet weight of 87.292% (15 318 quartets). The final bootstrap consensus tree (Fig. 4) recovered a strongly supported clade (BS = 100%) that contained all samples of P. gilesii, P. phylicifolia and the Evans Crown population, sister to P. scutellarioides. All samples of P. phylicifolia were recovered as a weakly supported clade (BS = 76%) sister to a strongly supported clade (BS = 100%) containing all samples of P. gilesii and the Evans Crown population. Within this clade, all samples of P. gilesii formed a strongly supported clade (BS = 100%) and all samples of the Evans Crown population formed a strongly supported clade (BS = 100%).
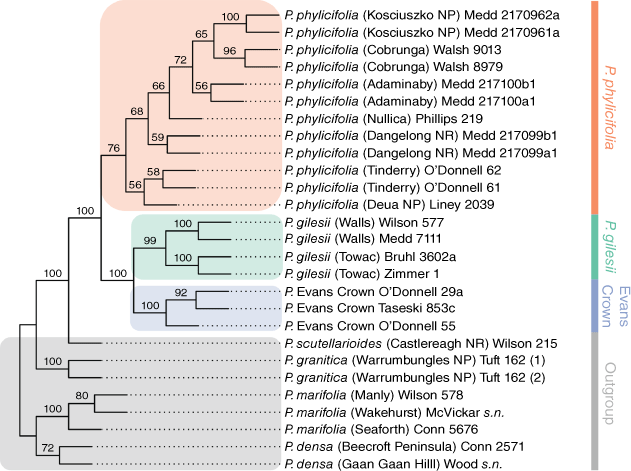
|
sNMF cluster analysis
On the basis of flattening of the cross-entropy curve (Supplementary Fig. S1), K = 3 was the most likely number of ancestral populations represented in the data. Estimates of ancestry coefficients for K = 3 recovered all samples of P. phylicifolia as one cluster, all samples of the Evans Crown population as a second cluster, and all samples of P. gilesii as a third cluster (Fig. 5). Each cluster exhibited little to no shared ancestry with other clusters. To compare alternative delimitation models, results for K = 3 were compared with results for K = 2 and K = 4. For K = 2; all samples of P. phylicifolia formed a cluster, and all samples of the Evans Crown population formed a cluster. Samples of P. gilesii were recovered as individuals with shared ancestry from both clusters. For K = 4, samples of P. phylicifolia were grouped into two clusters; the first comprised samples from Tinderry and Dangelong Nature Reserve, and the second comprised samples from Kosciuszko National Park, Cobrunga and Adaminaby. Further clustering was seen within P. phylicifolia for K ranging from 5 to 7. Separation of both populations of P. gilesii was seen upward of K = 6.
Population genetic analysis
Following removal of putative clones, the highest population-scale FST value of 0.651 was observed between the Evans Crown population and the Adaminaby population of P. phylicifolia (Supplementary Fig. S2). The lowest FST value (FST = 0.159) was between the Dangelong and Adaminaby populations of P. phylicifolia, but all pairwise FST estimates were significantly greater than 0 (P < 0.001; Fig. S2). Observed and expected heterozygosity were highest in the Dangelong population of P. phylicifolia (HO = 0.172; HE = 0.168) and lowest in the Evans Crown population (HO = 0.054; HE = 0.055; Tables 1, 2). At the species level, the highest FST value of 0.630 was observed between the Evans Crown population and P. gilesii, whereas the lowest FST value of 0.306 was observed between P. gilesii and P. phylicifolia (Supplementary Fig. S3).

|
Morphometric analysis
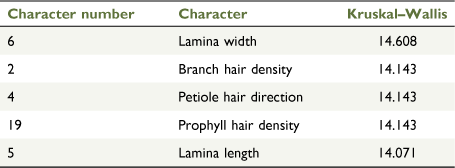
Cluster analysis recovered three groups at a dissimilarity value of 0.2233 (Fig. 6a). The first group consisted of all specimens of the Evans Crown population, the second consisted of all specimens of P. gilesii, and the third consisted of all specimens of P. phylicifolia. The top five characters that contributed to the separation of these groups on the basis of Kruskal–Wallis values were lamina width, branch hair density, petiole hair direction, prophyll hair density and lamina length (Table 3).
Three discrete clusters were observed in SSH-MDS ordination plots; one cluster contained all specimens of the Evans Crown population; another contained all samples of P. gilesii; and the remaining cluster contained all samples of P. phylicifolia (Fig. 6b). The stress value of 0.0503 is interpreted as low, suggesting that the ordination is an accurate representation of the dataset in reduced dimensionality (Belbin and Collins 2013). Petiole hair direction and prophyll hair density, which were shown to be informative characters on the basis of Kruskal–Wallis values in the cluster analysis, were also recovered with R2 values of >0.9 for the ordination (Supplementary Table S5).
Discussion
In this study, integration of genomic SNPs and morphometric analysis provided robust evidence in support of P. gilesii and the population from Evans Crown as distinct evolutionary lineages, and yielded insights into the population structure of the more widespread P. phylicifolia. O’Donnell et al. (2021a) first identified a close relationship among P. gilesii, P. phylicifolia and the Evans Crown population, but molecular data derived from Sanger sequencing were unable to satisfactorily resolve genetic boundaries among them. Here, analyses of genomic SNPs provide further support for the relationships recovered by O’Donnell et al. (2021a) (Fig. 4) and establish the presence of discrete genetic boundaries among the three taxa (Fig. 2, 3, 5, S2, S3). Our results demonstrated that they each form reciprocally monophyletic lineages that are strongly differentiated at many nuclear loci and morphologically distinguishable, and are thus capable of being recognised as independently evolving lineages and discrete taxonomic entities, i.e. species.
Ordination analyses of SNP data (Fig. 2) recovered three core clusters congruent with the two named and one putative species. Both the neighbour-net network graph (Fig. 3) and sNMF ancestry coefficient estimate plots (Fig. 5) demonstrated little reticulation or recombination among the three groups, indicating that gene flow between these groups is highly restricted. Although phylogenies recovered in this study (Fig. 4) and by O’Donnell et al. (2021a) both indicate a sister relationship between P. gilesii and the Evans Crown population, pairwise FST values between the two (Table 3, Fig. S3) suggest that they are more genetically differentiated from one another than either is from P. phylicifolia, despite the small geographic distance between P. gilesii and the Evans Crown population. Pronounced genetic differentiation among fragmented but geographically proximal populations and species has been similarly observed in other range-restricted granitic inselberg taxa (Hmeljevski et al. 2017; Bezemer et al. 2019; Robins et al. 2020). The observed heterozygosity values reported here (Tables 1, 2) suggest that accelerated genetic drift in historically small populations has likely contributed to the high FST values reported for P. gilesii and the Evans Crown population. The persistence of small populations is consistent with expectations for old, climatically buffered, infertile landscapes (OCBILs; Hopper et al. 2021). Nevertheless, the values reported here indicate substantial genetic divergence at the population and putative species level.

|
|
O’Donnell et al. (2021a) recovered two distinct clades of P. phylicifolia sens. str., namely, a ‘western’ clade comprising populations occurring along the Victorian Alps and Snowy Mountains, and an ‘eastern’ clade comprising populations from Tinderry south to Dangelong. The phylogeny estimated in this study (Fig. 4) placed all samples of P. phylicifolia within a clade but did not recover the same eastern and western groupings. Neighbour-network analysis (Fig. 3) recovered clusters with a membership similar to the eastern and western clades recovered by O’Donnell et al. (2021a), but both branches exhibited moderate amounts of reticulation. Morphological analyses (Fig. 6) did not detect substantial variation among populations of P. phylicifolia; however, clusters recovered within this group did exhibit some geographic partitioning. For example, specimens from Mount Coopracambra, Tinderry and Deua National Park formed a cluster corresponding with the eastern clade, whereas samples from Wulgulmerang, Adaminaby and Kosciuszko National Park clustered together, corresponding with the western clade. Specimens from Dangelong Nature Reserve and Tuross Falls formed an intermediate cluster between these groups.
Within Prostanthera, similar east–west divergences across the same geographic regions were observed in morphological, phytochemical, and molecular analyses of the P. lasianthos Labill. complex (Conn et al. 2021). Morphological and phytochemical evidence supported the presence of distinct east–west phenotypes, albeit with marginal phylogenetic differentiation. Subsequently, a western lineage of P. lasianthos (congruent in distribution to the western clade of P. phylicifolia) occurring on the Southern Tablelands, predominantly within Kosciuszko National Park, was segregated as the new species P. subalpina B.J.Conn & K.M.Proft. Geographically partitioned genetic structure has been similarly observed in multiple plant and animal groups across the Australian Alps (Osborne et al. 2000; Mitrovski et al. 2007; Koumoundouros et al. 2009; Slatyer et al. 2014; Endo et al. 2015; Haines et al. 2017; Bell et al. 2018; Atkins et al. 2020; Sumner et al. 2021; Umbers et al. 2022) and the eastern Tallaganda–Monaro regions (Garrick et al. 2004, 2007, 2012; Bull et al. 2013; Carlson et al. 2016). The genetic signatures recovered by these studies are consistent with what would be expected following repeated glaciation events, and suggest that Pleistocene climatic oscillations and repeated glaciation of the Kosciuszko Massif and Australian Alps may have driven genetic diversity and lineage divergence within this region (Byrne et al. 2011; Endo et al. 2015; Bell et al. 2018).
Because geographically partitioned lineages have been observed in other taxonomic groups across the Australian Alps and Monaro regions, it is possible that the eastern and western groups of P. phylicifolia represent lineages that are either in the process of diverging, or, conversely, lineages that were once geographically isolated that have become reconnected. On the basis of the phylogenetic placement of the Nullica population of P. phylicifolia between putative eastern and western groups, it is possible that this population represents a point of historical contact between two diverging lineages; however, this too is difficult to assess given the limited sampling of this population in this study. Few studies have investigated the Australian Alps and eastern Tallaganda–Monaro region in tandem, or the corridors connecting them. Of studies that surveyed both regions, some east–west genetic partitioning has been observed, although, in most cases, the geographic scale of these studies was too broad to distinguish this pattern with confidence (Searle et al. 2000; Chapple et al. 2005; Symula et al. 2008; Worth et al. 2011). Bell et al. (2018) emphasised that patterns of population connectivity across the Australian alpine flora have not been thoroughly interrogated, and are subsequently poorly understood.
Although two separate clades of P. phylicifolia s. str. were recovered by O’Donnell et al. (2021a), the presence of gene flow and lack of substantial morphological differentiation demonstrated here suggest that these clades are likely to be representative of a single taxon not warranting further subdivision. All populations share a cohesive morphology, which is congruent with Mueller’s protologue and type localities (von Mueller 1858). Our results therefore support the recognition of populations from the Victorian Alps and Snowy Mountains, Monaro, and Southern Tablelands of New South Wales as P. phylicifolia sens. str., as originally published by von Mueller (1858). The remaining taxonomic quandary, then, is the question of how to treat P. gilesii and the Evans Crown population. O’Donnell et al. (2021a) proposed the following three possible options: (1) subsume P. gilesii and the Evans Crown population into an enlarged P. phylicifolia; (2) retain P. phylicifolia and P. gilesii as distinct species with the Evans Crown population synonymised with the latter; or (3) recognise all three taxa as distinct species. Although P. phylicifolia, P. gilesii and the Evans Crown population are closely related, gene flow among them is highly restricted, and the phenetic differences detected in this study suggest that they are each morphologically discrete entities. To treat all three as synonymous would fail to recognise the genetic and morphological diversity present. Phylogenetic analyses place P. gilesii and the Evans Crown population as sister taxa; however, molecular evidence presented in this study suggests that the Evans Crown population is more genetically distinct from P. gilesii than it is from P. phylicifolia. Treating P. gilesii and the Evans Crown population as synonymous would fail to represent the relationship between them accurately. On the basis of the congruence of evidence presented here, the most defensible position is to treat all three taxa as independently evolving metapopulation lineages (sensu de Queiroz 2007) that warrant species-level recognition. The Evans Crown population is demonstrably genetically and morphologically distinct from P. gilesii and P. phylicifolia and is, consequently, described here as P. volucris R.P.O’Donnell. Prostanthera phylicifolia is also lectotypified here to clarify the application of this name, and to restrict its usage to populations in southern New South Wales and Victoria, as per the findings of O’Donnell et al. (2021a).
Taxonomy
Prostanthera phylicifolia F.Muell. Fragm. 1(1): 19 (1858)
Type citation: ‘In vertice rupestri montis McFarlane altitudine 4–5000’, nec non in rupibus secus rivulos districtus Maneroo.’ [‘At the summit of rocky mountain McFarlane from altitudes of 4–5000’, and also from cliffs or small rivulets in the district Maneroo’]. Type: Australia: Victoria: Eastern Highlands: ‘Mt M’Farlan’, s. dat., F. Mueller s.n. (lecto (here designated): K 000975463, left-hand side of sheet, right-hand sprig); isolecto: K 000975463, left-hand side of sheet, left-hand sprig; syn: MEL 43499 (right-hand side of sheet), MEL 43500).
Notes
There are several specimens of P. phylicifolia that are considered to be collections made by von Mueller (MEL 43499: left-hand side, MEL 43501, K 000975461); however, the handwriting on these respective accessions does not match that of Mueller’s and is subsequently of uncertain authorship. Because it is unclear whether these accessions were indeed collected and inspected by Mueller, they are not considered to comprise part of the original material of this name. Specimen K 000975462 is a collection from a locality described in Mueller’s handwriting as ‘Mitta Mitta’. This locality is not mentioned in Mueller’s protologue, and this specimen is therefore not part of the original material for this name.
Because O’Donnell et al. (2021a) demonstrated that P. phylicifolia sens. str. is restricted to southern New South Wales and Victoria, Mueller’s locality description of ‘Maneroo’ must be considered to be an early spelling of Monaro (referring to the Monaro region), and not the rural locality of Maneroo located in Longreach, Queensland. Although specimens bearing the locality description ‘Maneroo’ (K000975460, K000975461) appear to be conspecific with the Mount McFarlane specimens, they have been excluded as syntypes because of the uncertain application of this locality name.
Specimen K000975461 bears a label of uncertain authorship reading ‘var. velutina’, and K000975462 is similarly labelled with ‘v. velutina’, albeit in this instance written definitively in Mueller’s handwriting. Although Mueller’s protologue describes individuals of P. phylicifolia as ‘glabra v. velutina’ (i.e. glabrous or velutinous; von Mueller 1858, p. 19), neither of these qualifiers are separated by Mueller as named varieties. Therefore, the name P. phylicifolia var. velutina appears to be a manuscript name of no nomenclatural standing that has never been published. O’Donnell et al. (2021a) found little genetic differentiation among individuals of P. phylicifolia that were predominantly glabrous and individuals that were densely hairy. Because most specimens of P. phylicifolia examined in this study were predominantly glabrous and some of the glabrous syntypes were well endowed with flowers and seemingly fruiting calyces, a representatively glabrous specimen is designated as the lectotype.
Type: Australia: New South Wales: Central Tablelands: Evans Crown Nature Reserve, ~2.8 km SE of Tarana township, 28 Oct. 2018, G.M. Taseski 853, (holo: NSW 1055966 (sheet) and NSW 1057497 (spirit; listed as an associated collection on NSW 1055966); iso: BRI, CANB, K, MEL, MO, NE 110628, P, UNSW). Prostanthera sp. Evans Crown (G.M.Taseski NSW 1055966) (O’Donnell et al. 2021a).

|
Diagnosis
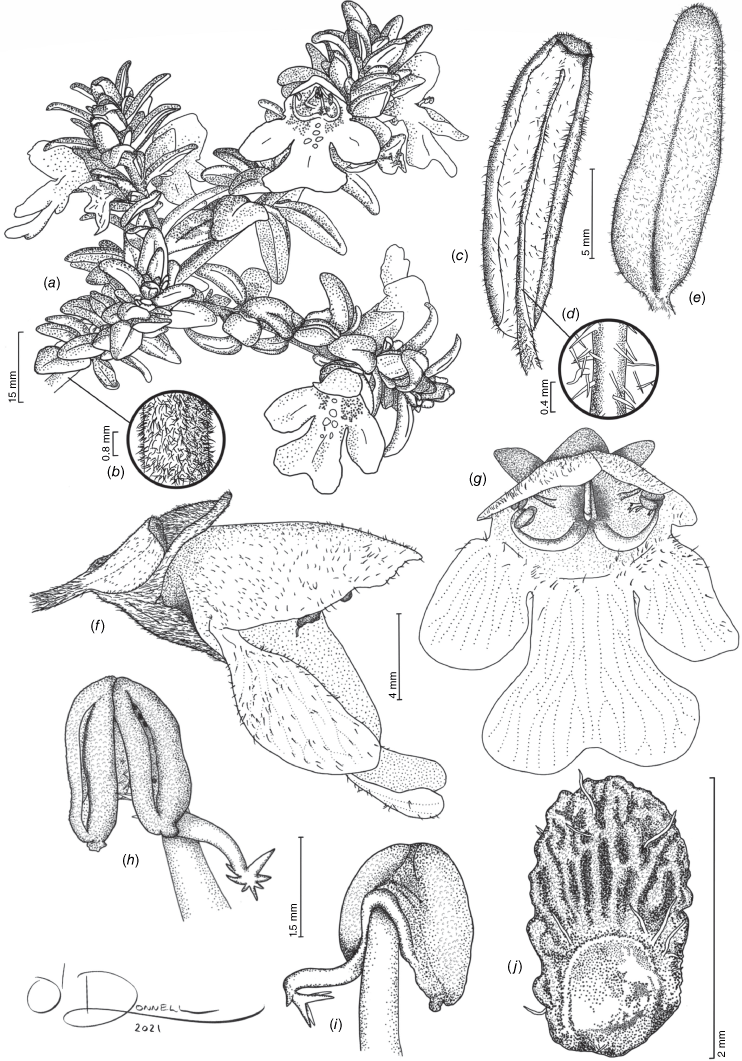
Prostanthera volucris can be distinguished from the morphologically similar P. gilesii by its larger prophylls, 3.8–9 mm long, 0.8–4.5 mm wide (v. <4 mm long, <0.6 mm wide for P. gilesii), densely hairy branches, calyces and prophylls (up to 80 v. up to 40 trichomes mm−2 for P. gilesii), appressed to subappressed retrorse trichomes (v. antrorse for P. gilesii) and densely hairy abaxial and adaxial lamina surfaces (v. predominantly glabrous with occasional antrorse trichomes restricted to the abaxial surface midrib for P. gilesii). Prostanthera volucris can also be distinguished from P. phylicifolia and P. gilesii by its mericarps that are rugose and papillose, with occasional long pilose trichomes (v. reticulate, not distinctly papillose and glabrous for P. phylicifolia; mature mericarps have never been observed for P. gilesii; Table 4).

|
Compact, erect shrub up to 80 cm high, with plants forming tight mats. Branches ± terete, occasionally quadrangular early in development, becoming terete with age, densely hairy (20–80 trichomes mm−2); trichomes 0.1–0.4 mm long, appressed to subappressed, retrorse, straight to slightly curled, white; sessile glands indistinct or absent (obscured by indumentum). Leaves velutinous, appearing silver–light green, slightly paler on abaxial surface, occasionally becoming red after prolonged exposure to strong sunlight, not aromatic when touched or crushed; petiole 0.88–1.4 mm long, densely hairy (20–48 trichomes mm−2), the trichomes 0.2–0.8 mm long, appressed to subappressed, retrorse, straight to slightly curled, white; lamina narrowly ovate to elliptic, 12–19 mm long, 3–5 mm wide, with length to width ratio 2.6–5.3 and length of maximum width from base to total lamina length ratio 0.1–0.4; abaxial surface moderately to densely hairy, particularly on midrib (16–40 trichomes mm−2), the trichomes 0.2–0.4 mm long, spreading to erect (occasionally retrorse along midrib), straight to slightly curled, white; abaxial lamina glands indistinct; adaxial surface moderately to densely hairy (16–30 trichomes mm−2), the trichomes 0.1–0.3 mm long, spreading to erect, straight to slightly curled, white; adaxial lamina glands indistinct; base obtuse (occasionally appearing attenuate because margin more involute towards base); margin recurved; apex obtuse; venation indistinct, the midrib slightly raised on the abaxial surface. Inflorescence a frondose botryoidal conflorescence of monadic uniflorescences with 4–8-flowers per conflorescence. Podium a1 axis 0.9–2.8 mm long, densely hairy (30–60[–88] trichomes mm−2), the trichomes 0.1–0.2 mm long, appressed to subappressed, retrorse, straight to slightly curled, white, with glands indistinct or absent (obscured by indumentum); anthopodium absent or indistinct. Pherophylls not seen. Prophylls ± persistent, inserted near base of calyx, opposite, narrow-ovate to narrowly elliptic, 3.8–9 mm long, 0.8–4.5 mm wide, the length to width ratio 1.8–5.6, the length of maximum width from base to total lamina length ratio 0.3–0.9, densely hairy (16–56 trichomes mm−2), the trichomes 0.1–0.2 mm long, appressed to subappressed, retrorse, straight to slightly curled, white, with glands indistinct or absent (obscured by indumentum); base slightly attenuate; margin entire; apex obtuse; venation indistinct. Calyx tube 1.5–2.1 mm long, light green, occasionally darkening to dark mauve with sun exposure; abaxial lobe ovate to broadly ovate, 2–4 mm long, 2–4 mm wide at base, the apex rounded to retuse; adaxial lobe ovate to elliptic, 3–5.3 mm long, 2.1–4.2 mm wide at base, the length to width ratio 1.2–1.4, the apex ± rounded; the adaxial lobe length to abaxial lobe length ratio ~1.3; outer surface densely hairy (24–64 trichomes mm−2), the trichomes 0.2–0.35 mm long, appressed to subappressed, retrorse, straight to slightly curled, white, with glands indistinct or absent (obscured by indumentum); inner surface of tube glabrous; inner surface of lobes moderately to densely hairy near margin and apex, the trichomes appressed to subappressed, spreading to occasionally antrorse, straight to slightly curled, white. Corolla 14–18 mm long, white, with purple to dark mauve speckled markings on the inner surface of the tube and pale orange to yellow markings on base of abaxial median lobe; tube 3–5 mm long; abaxial median lobe broadly spathulate, 7–9.5 mm long, 4–7 mm wide (below distal lobing), length to width ratio 1.4–1.8, the apex slightly irregular and rounded, bilobed (sinus 2–2.3 mm long, 2.5–3 mm wide distally); lateral lobes oblong to slightly elliptic, 5.4–5.7 mm long, 3.1–3.8 mm wide, length to width ratio 1.5–1.8, the apex rounded, slightly irregular; adaxial median lobe-pair broad to depressed-ovate, 6–7 mm long, 9–9.5 mm wide, length to width ratio 0.6–0.8, the apex rounded, irregular, bilobed (sinus 0.6–0.8 mm long, 0.9–1.6 mm wide, the median margin of lobes occasionally overlapping slightly); outer surface sparsely glandular, moderately hairy, particularly on lobes (8–24 trichomes mm−2), the trichomes 0.1–0.4 mm long, erect to spreading along tube before becoming appressed to subappressed and antrorse on lobes; inner surface ± glabrous, the lobes sparsely hairy to moderately hairy at the tube opening rim and sinuses between lobes, the trichomes 0.1–0.2 mm long, crinkled. Stamens inserted 2.8–4.1 mm above base of corolla; filaments 3.1–5.4 mm long, white, often with mauve tinge; anthers 1.4–1.8 mm long, minutely papillose with an acumen at the base of each lobe and trichomes between lobes, the trichomes 0.1–0.2 mm long, white; connective appendage 0.8–1.5 mm long, dark mauve maturing to yellowish-brown, with a few narrowly triangular trichomes that are 0.1–0.35 mm long and white. Disc 0.5–1 mm long. Pistil 7.5–9.5 mm long; ovary cylindrical-obovoid, 0.9–1 mm long, at base 0.9–1 mm diameter, the lobes 0.5–0.65 mm long; style 7.5–8 mm long; stigma lobes 0.3–0.4 mm long. Fruiting calyx not strongly accrescent, the abaxial lobe clasping to conceal developing mericarps, the adaxial lobe not strongly reflexed. Mature mericarps 1.8–2.1 mm long, 1–1.2 mm wide, rugose, minutely papillose, with a few spreading trichomes that are 0.2–0.5 mm long and white.
Distribution
Known from a single granitic tor in the Evans Crown Nature Reserve, south-east of Tarana, New South Wales, Australia (Fig. 1). This location is situated within the Central Tablelands Botanical Division and South Eastern Highlands IBRA Region (NSW National Parks and Wildlife Service 2009).
Habitat
Prostanthera volucris grows on exposed granite formations at 1000–1020 m altitude, along drainage crevices in shallow, skeletal humic soils, with Cyphanthera albicans and Cheilanthes sieberi nearby.
Etymology
From the Latin ‘volucris’ (‘winged’ or ‘winged creature’), in reference to the substantial and densely hairy prophylls, which, inserted at the base of the calyx, give the calyx the appearance of being winged.
Ecology and conservation
Although the first collector of P. volucris described it as ‘locally abundant in rock crevices’ (McKee 7043, NSW 237164), only one population is known. More intensive surveys of the Evans Crown Nature Reserve and other unexplored vegetation islands in the nearby area should be undertaken. Because P. volucris is known to grow only on exposed, granitic outcrops, future attempts to locate additional populations should prioritise these landscape features.
The pollinators of P. volucris are unknown; however, its floral characteristics correspond with a floral type visited primarily by bees, although not excluding other insects such as flies (Wilson et al. 2017). Studies of foraging ranges in Australian bees found a typical maximum foraging range of ~700 m (Smith et al. 2017), which suggests that pollen is unlikely to travel further than this distance. Because P. volucris grows in tight, tangled mats, it is difficult to determine the limits of individual plants. Pairwise shared allele frequencies indicate that some samples represent ramets from a clonal parent, whereas others represent separate individuals. Seedlings have been observed in recent surveys and mature individuals have been observed to set seed; however, it is unknown what proportion of seed set is viable. Because P. volucris is known to occur along drainage crevices, it is likely that seeds are being dispersed primarily by water runoff.
It is unknown whether P. volucris is subject to herbivory; however, as other species of Prostanthera have been observed to be grazed by herbivores, similar threats may apply in this instance (Jusaitis 2018). Sheep and goats have been known to access the reserve from neighbouring properties and there are currently no feral animal control programs implemented for the reserve (NSW National Parks and Wildlife Service 2009). The reserve is a popular attraction for hikers and rock climbers, and because the species occurs on a geologically striking outcrop, human activity is a likely threat to this population. On revisiting the site following the severe drought conditions of 2019–2020, the population appeared to be severely affected by heat and drought stress and had declined substantially in condition (G. M. Taseski, pers. obs., 2020). Projected prolonged drought conditions and heightened temperatures may adversely affect this population, particularly on account of its highly exposed habitat.
Given the horticultural potential of P. volucris, this species could be a target of increased collecting pressure. Attempts are underway to develop an ex situ collection with botanic gardens and native plant wholesalers. The known distribution of this species is restricted such that the area of occupancy and extent of occurrence are not greater than 4 km2. Because this species is known from only one highly restricted population of <250 mature individuals where decline in response to drought stress has been observed, we suggest that this species satisfies the criteria to be considered Critically Endangered under the New South Wales Biodiversity Conservation Act 2016 (see https://www.legislation.nsw.gov.au/view/html/inforce/current/act-2016-063#statusinformation), the Environment Protection and Biodiversity Conservation Act 1999 and the IUCN Red List criteria thresholds (International Union for Conservation of Nature and Natural Resources 2022).
Notes
Although quantifiable mericarp characters were not included for morphological analyses, differences in mericarp morphology were observed between P. volucris and P. phylicifolia. The mericarp is rugose and distinctly papillate in P. volucris, whereas it is reticulate and not distinctly papillose in P. phylicifolia. Scant attention has been paid to mericarp surface ornamentation and sculpting in recent descriptions of Prostanthera, with the exception of Williams et al. (2006) in their treatment of the P. spinosa complex and Guerin’s (2005) study of mericarp morphology in the Westringieae. The findings outlined here and by Guerin (2005) and Williams et al. (2006) highlight that mericarp morphology may be taxonomically informative in Prostanthera. Further examination of mericarp morphology across the genus is warranted and may provide additional characters for distinguishing among closely related species in future diagnoses.
Other specimens examined
AUSTRALIA: NEW SOUTH WALES: CENTRAL TABLELANDS: South Eastern Highlands: Evans Crown Nature Reserve: H.S. McKee 7043, 10 Jan. 1960 (NSW237164); A.N. Rodd 11009, 9 Mar. 2002 (NSW856887); R.P. O’Donnell & G.M. Taseski 28, 7 Apr. 2020 (NSW1100357); R.P. O’Donnell & G.M. Taseski 29, 7 Apr. 2020 (NSW1100369); R.P. O’Donnell & G.M. Taseski 30, 7 Apr. 2020 (NSW1100379); R.P. O’Donnell & T.C. Wilson 55, 4 Oct. 2020 (NSW1100402); R.P. O’Donnell & T.C. Wilson 56, 4 Oct. 2020 (NSW1100403).
Supplementary material
Supplementary material is available online.
Data availability
All R code used in this study is available at https://github.com/rpodonnell/ASB_PEC. SNP data and metadata are available at the Research UNE data repository at https://hdl.handle.net/1959.11/53809. This paper was first made available as a preprint (O’Donnell et al. 2021b).
Conflicts of interest
Jeremy J. Bruhl is an Associate Editor of Australian Systematic Botany but did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Declaration of funding
Ryan P. O’Donnell was supported by an N.C.W. Beadle Herbarium Botany Scholarship and the School of Environmental and Rural Science (SERS) at the University of New England. This project received support from the New South Wales Government’s Saving our Species program under the Department of Planning, Industry and Environment (DPIE). Trevor C. Wilson was supported by the Australian Biological Resources Study (ABRS) grant (RG19-17).
Acknowledgements
We thank the National Herbarium of New South Wales (NSW) and the N.C.W. Beadle Herbarium (NE) for access to collections; the Research Centre for Ecosystem Resilience (Royal Botanic Gardens and Domain Trust) for sharing DArTseq data; Richard Medd for collections of P. gilesii and P. phylicifolia and logistics; Neville Walsh (MEL) for silica-gel material from Victorian populations of P. phylicifolia; Danielle Smith and Helen Kennedy (NE) for sample preparation; Lesley Elkan (NSW) for constructive feedback on illustrations; Ben Correy and Shane Smith (NSW National Parks and Wildlife Service) for correspondence and logistics; Markus Buchhorn for access to and assistance on his property; and Benjamin Anderson (PERTH), Stephen Hopper (UWA), Katharina Nargar (CNS) and Kevin Thiele (UWA) for constructive feedback on the manuscript.
References
Atkins ZS, Amor MD, Clemann N, Chapple DG, While GM, Gardner MG, Haines ML, Harrisson KA, Schroder M, Robert KA (2020) Allopatric divergence drives the genetic structuring of an endangered alpine endemic lizard with a sky-island distribution. Animal Conservation 23, 104–118.| Allopatric divergence drives the genetic structuring of an endangered alpine endemic lizard with a sky-island distribution.Crossref | GoogleScholarGoogle Scholar |
Australian Plant Census (2021) APC: Prostanthera. In ‘The Australian National Species List: Vascular Plants’. APC (version 51411005). (Council of Heads of Australasian Herbaria) Available at https://biodiversity.org.au/nsl/services/rest/treeElement/51411005/51380123 [Verified 9 January 2023]
Australia’s Virtual Herbarium (2021) Occurrence records: Prostanthera phylicifolia, P. gilesii and P. sp. Evans Crown. In ‘The Australasian Virtual Herbarium’. (Atlas of Living Australia) Available at https://avh.ala.org.au/occurrences/search?q=%28taxa%3A%22Prostanthera+phylicifolia%22+OR+taxa%3A%22Prostanthera+gilesii%22+OR+raw_scientificName%3A%22Prostanthera+sp.+Evans+Crown%22%29#tab_mapView [Verified 9 January 2023]
Bean AR (2014) Four new Queensland species of Solanum L. allied to S. ellipticum R.Br. (Solanaceae). Austrobaileya 9, 216–228.
Belbin L, Collins A (2013) ‘PATN - Finding Patterns in Data.’ (Blatant Fabrications)
Bell N, Griffin PC, Hoffmann AA, Miller AD (2018) Spatial patterns of genetic diversity among Australian alpine flora communities revealed by comparative phylogenomics. Journal of Biogeography 45, 177–189.
| Spatial patterns of genetic diversity among Australian alpine flora communities revealed by comparative phylogenomics.Crossref | GoogleScholarGoogle Scholar |
Bentham G (1870) Labiatae. In ‘Flora Australiensis. Vol. 5’. (Eds G Bentham, F Mueller) pp. 70–137. (Reeve: London, UK)
Bezemer N, Krauss SL, Roberts DG, Hopper SD (2019) Conservation of old individual trees and small populations is integral to maintain species’ genetic diversity of a historically fragmented woody perennial. Molecular Ecology 28, 3339–3357.
| Conservation of old individual trees and small populations is integral to maintain species’ genetic diversity of a historically fragmented woody perennial.Crossref | GoogleScholarGoogle Scholar |
Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution 21, 255–265.
| Neighbor-net: an agglomerative method for the construction of phylogenetic networks.Crossref | GoogleScholarGoogle Scholar |
Bull JK, Sands CJ, Garrick RC, Gardner MG, Tait NN, Briscoe DA, Rowell DM, Sunnucks P (2013) Environmental complexity and biodiversity: the multi-layered evolutionary history of a log-dwelling velvet worm in montane temperate Australia. PLoS One 8, e84559
| Environmental complexity and biodiversity: the multi-layered evolutionary history of a log-dwelling velvet worm in montane temperate Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Steane DA, Joseph L, Yeates DK, Jordan GJ, Crayn D, Aplin K, Cantrill DJ, Cook LG, Crisp MD, Keogh JS, Melville J, Moritz C, Porch N, Sniderman JMK, Sunnucks P, Weston PH (2011) Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. Journal of Biogeography 38, 1635–1656.
| Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota.Crossref | GoogleScholarGoogle Scholar |
Carlson E, MacDonald AJ, Adamack A, McGrath T, Doucette LI, Osborne WS, Gruber B, Sarre SD (2016) How many conservation units are there for the endangered grassland earless dragons? Conservation Genetics 17, 761–774.
| How many conservation units are there for the endangered grassland earless dragons?Crossref | GoogleScholarGoogle Scholar |
Chappill JA, Ladiges PY (1992) Geographical variation in morphology of Eucalyptus mannifera and allied taxa. Australian Systematic Botany 5, 359–372.
| Geographical variation in morphology of Eucalyptus mannifera and allied taxa.Crossref | GoogleScholarGoogle Scholar |
Chapple DG, Keogh JS, Hutchinson MN (2005) Substantial genetic substructuring in southeastern and alpine Australia revealed by molecular phylogeography of the Egernia whitii (Lacertilia: Scincidae) species group. Molecular Ecology 14, 1279–1292.
| Substantial genetic substructuring in southeastern and alpine Australia revealed by molecular phylogeography of the Egernia whitii (Lacertilia: Scincidae) species group.Crossref | GoogleScholarGoogle Scholar |
Chifman J, Kubatko L (2014) Quartet inference from SNP data under the coalescent model. Bioinformatics 30, 3317–3324.
| Quartet inference from SNP data under the coalescent model.Crossref | GoogleScholarGoogle Scholar |
Chifman J, Kubatko L (2015) Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites. Journal of Theoretical Biology 374, 35–47.
| Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites.Crossref | GoogleScholarGoogle Scholar |
Collins TL, Schmidt-Lebuhn AN, Andrew RL, Telford IRH, Bruhl JJ (2022) There’s gold in them thar hills! Morphology and molecules delimit species in Xerochrysum (Asteraceae; Gnaphalieae) and reveal many new taxa. Australian Systematic Botany 35, 120–185.
| There’s gold in them thar hills! Morphology and molecules delimit species in Xerochrysum (Asteraceae; Gnaphalieae) and reveal many new taxa.Crossref | GoogleScholarGoogle Scholar |
Conn BJ (1984) A taxonomic revision of Prostanthera Labill. section Klanderia (F.v. Muell.) Benth. (Labiatae). Journal of the Adelaide Botanic Gardens 6, 207–348.
Conn BJ (1988) A taxonomic revision of Prostanthera Labill. section Prostanthera (Labiatae), 1. The species of the Northern Territory, South Australia and Western Australia. Nuytsia 6, 351–411.
Conn BJ, Wilson TC (2015) Two new species of Prostanthera (Lamiaceae) in New South Wales. Telopea 18, 463–474.
| Two new species of Prostanthera (Lamiaceae) in New South Wales.Crossref | GoogleScholarGoogle Scholar |
Conn BJ, Wilson TC, Henwood MJ, Proft K (2013) Circumscription and phylogenetic relationships of Prostanthera densa and P. marifolia (Lamiaceae). Telopea 15, 149–164.
| Circumscription and phylogenetic relationships of Prostanthera densa and P. marifolia (Lamiaceae).Crossref | GoogleScholarGoogle Scholar |
Conn BJ, Henwood MJ, Proft K, Wilson TC (2016) Molecular phylogenetics reveals a new species of Prostanthera from tropical Queensland with links to more southerly taxa. Telopea 19, 13–22.
| Molecular phylogenetics reveals a new species of Prostanthera from tropical Queensland with links to more southerly taxa.Crossref | GoogleScholarGoogle Scholar |
Conn BJ, Henwood MJ, Proft KM, Scott JA, Wilson TC, Howes RS (2021) An integrative taxonomic approach resolves the Prostanthera lasianthos (Lamiaceae) species complex. Australian Systematic Botany 34, 438–476.
| An integrative taxonomic approach resolves the Prostanthera lasianthos (Lamiaceae) species complex.Crossref | GoogleScholarGoogle Scholar |
Copeland LM, Bruhl JJ, Craven LA, Brubaker CL (2007) Phenetic analyses of Homoranthus (Myrtaceae: Chamelaucieae) on the basis of morphology. Australian Systematic Botany 20, 417–427.
| Phenetic analyses of Homoranthus (Myrtaceae: Chamelaucieae) on the basis of morphology.Crossref | GoogleScholarGoogle Scholar |
Cruz VMV, Kilian A, Dierig DA (2013) Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop Lesquerella and related species. PLoS One 8, e64062
| Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop Lesquerella and related species.Crossref | GoogleScholarGoogle Scholar |
Dayrat B (2005) Towards integrative taxonomy. Biological Journal of the Linnean Society 85, 407–415.
| Towards integrative taxonomy.Crossref | GoogleScholarGoogle Scholar |
de Queiroz K (2007) Species concepts and species delimitation. Systematic Biology 56, 879–886.
| Species concepts and species delimitation.Crossref | GoogleScholarGoogle Scholar |
de Salas MF, Schmidt-Lebuhn AN (2018) Integrative approach resolves the taxonomy of the Ozothamnus ledifolius (Asteraceae: Gnaphaliae) species complex in Tasmania, Australia. Phytotaxa 358, 117–138.
| Integrative approach resolves the taxonomy of the Ozothamnus ledifolius (Asteraceae: Gnaphaliae) species complex in Tasmania, Australia.Crossref | GoogleScholarGoogle Scholar |
Endo Y, Nash M, Hoffmann AA, Slatyer R, Miller AD (2015) Comparative phylogeography of alpine invertebrates indicates deep lineage diversification and historical refugia in the Australian Alps. Journal of Biogeography 42, 89–102.
| Comparative phylogeography of alpine invertebrates indicates deep lineage diversification and historical refugia in the Australian Alps.Crossref | GoogleScholarGoogle Scholar |
Fernández-Mazuecos M, Mellers G, Vigalondo B, Sáez L, Vargas P, Glover BJ (2017) Resolving recent plant radiations: power and robustness of genotyping-by-sequencing. Systematic Biology 67, 250–268.
| Resolving recent plant radiations: power and robustness of genotyping-by-sequencing.Crossref | GoogleScholarGoogle Scholar |
Fitzsimons JA, Michael DR (2017) Rocky outcrops: a hard road in the conservation of critical habitats. Biological Conservation 211, 36–44.
| Rocky outcrops: a hard road in the conservation of critical habitats.Crossref | GoogleScholarGoogle Scholar |
Freeland JR, Petersen S, Kirk H (2011) ‘Molecular Ecology.’ (Wiley-Blackwell: Hoboken, NJ, USA)
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods in Ecology and Evolution 6, 925–929.
| LEA: an R package for landscape and ecological association studies.Crossref | GoogleScholarGoogle Scholar |
Garrick RC, Sands CJ, Rowell DM, Tait NN, Greenslade P, Sunnucks P (2004) Phylogeography recapitulates topography: very fine-scale local endemism of a saproxylic ‘giant’ springtail at Tallaganda in the Great Dividing Range of south-east Australia. Molecular Ecology 13, 3329–3344.
| Phylogeography recapitulates topography: very fine-scale local endemism of a saproxylic ‘giant’ springtail at Tallaganda in the Great Dividing Range of south-east Australia.Crossref | GoogleScholarGoogle Scholar |
Garrick RC, Sands CJ, Rowell DM, Hillis DM, Sunnucks P (2007) Catchments catch all: long-term population history of a giant springtail from the southeast Australian highlands – a multigene approach. Molecular Ecology 16, 1865–1882.
| Catchments catch all: long-term population history of a giant springtail from the southeast Australian highlands – a multigene approach.Crossref | GoogleScholarGoogle Scholar |
Garrick RC, Rowell DM, Sunnucks P (2012) Phylogeography of saproxylic and forest floor invertebrates from Tallaganda, south-eastern Australia. Insects 3, 270–294.
| Phylogeography of saproxylic and forest floor invertebrates from Tallaganda, south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Georges A, Gruber B, Pauly GB, White D, Adams M, Young MJ, Kilian A, Zhang X, Shaffer HB, Unmack PJ (2018) Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia. Molecular Ecology 27, 5195–5213.
| Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Gruber B, Unmack PJ, Berry OF, Georges A (2018) DARTR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources 18, 691–699.
| DARTR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing.Crossref | GoogleScholarGoogle Scholar |
Guerin GR (2005) Nutlet morphology in Hemigenia R. Br. and Microcorys R. Br. (Lamiaceae). Plant Systematics and Evolution 254, 49–68.
| Nutlet morphology in Hemigenia R. Br. and Microcorys R. Br. (Lamiaceae).Crossref | GoogleScholarGoogle Scholar |
Haines ML, Stuart-Fox D, Sumner J, Clemann N, Chapple DG, Melville J (2017) A complex history of introgression and vicariance in a threatened montane skink (Pseudemoia cryodroma) across an Australian sky island system. Conservation Genetics 18, 939–950.
| A complex history of introgression and vicariance in a threatened montane skink (Pseudemoia cryodroma) across an Australian sky island system.Crossref | GoogleScholarGoogle Scholar |
Hmeljevski KV, Nazareno AG, Leandro Bueno M, dos Reis MS, Forzza RC (2017) Do plant populations on distinct inselbergs talk to each other? A case study of genetic connectivity of a bromeliad species in an Ocbil landscape. Ecology and Evolution 7, 4704–4716.
| Do plant populations on distinct inselbergs talk to each other? A case study of genetic connectivity of a bromeliad species in an Ocbil landscape.Crossref | GoogleScholarGoogle Scholar |
Hopper SD, Lambers H, Silveira FAO, Fiedler PL (2021) OCBIL theory examined: reassessing evolution, ecology and conservation in the world’s ancient, climatically buffered and infertile landscapes. Biological Journal of the Linnean Society 133, 266–296.
| OCBIL theory examined: reassessing evolution, ecology and conservation in the world’s ancient, climatically buffered and infertile landscapes.Crossref | GoogleScholarGoogle Scholar |
Hundsdoerfer AK, Lee KM, Kitching IJ, Mutanen M (2019) Genome-wide SNP data reveal an overestimation of species diversity in a group of hawkmoths. Genome Biology and Evolution 11, 2136–2150.
| Genome-wide SNP data reveal an overestimation of species diversity in a group of hawkmoths.Crossref | GoogleScholarGoogle Scholar |
Huson DH (1998) SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73.
| SplitsTree: analyzing and visualizing evolutionary data.Crossref | GoogleScholarGoogle Scholar |
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23, 254–267.
| Application of phylogenetic networks in evolutionary studies.Crossref | GoogleScholarGoogle Scholar |
International Union for Conservation of Nature and Natural Resources (2022) Guidelines for Using the IUCN Red List Categories and Criteria. Version 15.1 (July 2022). Prepared by the Standards and Petitions Committee of the IUCN Species Survival Commission. Available at https://www.iucnredlist.org/resources/redlistguidelines [Verified 9 January 2023]
Joyce EM, Pannell CM, Rossetto M, Yap J-YS, Thiele KR, Wilson PD, Crayn DM (2021) Molecular phylogeography reveals two geographically and temporally separated floristic exchange tracks between Southeast Asia and northern Australia. Journal of Biogeography 48, 1213–1227.
| Molecular phylogeography reveals two geographically and temporally separated floristic exchange tracks between Southeast Asia and northern Australia.Crossref | GoogleScholarGoogle Scholar |
Jusaitis M (2018) Threatened plant translocation case study: Prostanthera eurybioides (Monarto Mintbush), Lamiaceae. Australasian Plant Conservation 26, 15–17.
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. Methods in Molecular Biology 888, 67–89.
| Diversity arrays technology: a generic genome profiling technology on open platforms.Crossref | GoogleScholarGoogle Scholar |
Koumoundouros T, Sumner J, Clemann N, Stuart-Fox D (2009) Current genetic isolation and fragmentation contrasts with historical connectivity in an alpine lizard (Cyclodomorphus praealtus) threatened by climate change. Biological Conservation 142, 992–1002.
| Current genetic isolation and fragmentation contrasts with historical connectivity in an alpine lizard (Cyclodomorphus praealtus) threatened by climate change.Crossref | GoogleScholarGoogle Scholar |
Mijangos JL, Gruber B, Berry O, Pacioni C, Georges A (2022) dartR v2: an accessible genetic analysis platform for conservation, ecology and agriculture. Methods in Ecology and Evolution 13, 2150–2158.
| dartR v2: an accessible genetic analysis platform for conservation, ecology and agriculture.Crossref | GoogleScholarGoogle Scholar |
Minchin PR (1987) An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 69, 89–107.
| An evaluation of the relative robustness of techniques for ecological ordination.Crossref | GoogleScholarGoogle Scholar |
Mitrovski P, Heinze DA, Broome L, Hoffmann AA, Weeks AR (2007) High levels of variation despite genetic fragmentation in populations of the endangered mountain pygmy-possum, Burramys parvus, in alpine Australia. Molecular Ecology 16, 75–87.
| High levels of variation despite genetic fragmentation in populations of the endangered mountain pygmy-possum, Burramys parvus, in alpine Australia.Crossref | GoogleScholarGoogle Scholar |
Montalvo AM, Conard SG, Conkle MT, Hodgskiss PD (1997) Population structure, genetic diversity, and clone formation in Quercus chrysolepis (Fagaceae). American Journal of Botany 84, 1553–1564.
| Population structure, genetic diversity, and clone formation in Quercus chrysolepis (Fagaceae).Crossref | GoogleScholarGoogle Scholar |
Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA (2013) Genotyping-by-sequencing in ecological and conservation genomics. Molecular Ecology 22, 2841–2847.
| Genotyping-by-sequencing in ecological and conservation genomics.Crossref | GoogleScholarGoogle Scholar |
NSW Department of Planning and Environment (2019) Saving our species Prostanthera gilesii 2018-2019 annual report card. Available at https://www.environment.nsw.gov.au/-/media/OEH/Corporate-Site/Documents/Animals-and-plants/Threatened-species/Report-cards/2018-2019/02-site-managed-flora/prostanthera-gilesii-2018-19.PDF
NSW National Parks and Wildlife Service (2009) Evans Crown Nature Reserve Plan of Management. DECC 2009/140. (NSW Department of Environment and Climate Change: Australia) Available at https://www.environment.nsw.gov.au/-/media/OEH/Corporate-Site/Documents/Parks-reserves-and-protected-areas/Parks-plans-of-management/evans-crown-nature-reserve-plan-of-management-090140.pdf [Verified 9 January 2023]
O’Donnell RP, Wilson TC, Andrew RL, Telford IR, Taseski GM, Zimmer H, Bruhl JJ (2021a) Molecular phylogenetic analysis of the Prostanthera phylicifolia (Lamiaceae) assemblage resolves relationships of the ‘Critically Endangered’ P. gilesii and other putative new species. Telopea 24, 359–375.
| Molecular phylogenetic analysis of the Prostanthera phylicifolia (Lamiaceae) assemblage resolves relationships of the ‘Critically Endangered’ P. gilesii and other putative new species.Crossref | GoogleScholarGoogle Scholar |
O’Donnell RP, Bruhl JJ, Telford IRH, Wilson TC, Zimmer HC, Taseski GM, Andrew RL (2021b) Molecular and morphological analyses support recognition of Prostanthera volucris (Lamiaceae), a new species from the Central Tablelands of New South Wales. bioRxiv 2021.12.21.473648
| Molecular and morphological analyses support recognition of Prostanthera volucris (Lamiaceae), a new species from the Central Tablelands of New South Wales.Crossref | GoogleScholarGoogle Scholar | [Published online 22 December 2021]
Osborne MJ, Norman JA, Christidis L, Murray ND (2000) Genetic distinctness of isolated populations of an endangered marsupial, the mountain pygmy-possum, Burramys parvus. Molecular Ecology 9, 609–613.
| Genetic distinctness of isolated populations of an endangered marsupial, the mountain pygmy-possum, Burramys parvus.Crossref | GoogleScholarGoogle Scholar |
Padial JM, Miralles A, De la Riva I, Vences M (2010) The integrative future of taxonomy. Frontiers in Zoology 7, 16
| The integrative future of taxonomy.Crossref | GoogleScholarGoogle Scholar |
Pembleton LW, Cogan NOI, Forster JW (2013) StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Molecular Ecology Resources 13, 946–952.
| StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations.Crossref | GoogleScholarGoogle Scholar |
Plunkett GT, Bruhl JJ, Telford IRH (2009) Two new, sympatric species of Wahlenbergia (Campanulaceae) from the New England Tableland escarpment, New South Wales, Australia. Australian Systematic Botany 22, 319–331.
| Two new, sympatric species of Wahlenbergia (Campanulaceae) from the New England Tableland escarpment, New South Wales, Australia.Crossref | GoogleScholarGoogle Scholar |
Robins TP, Binks RM, Byrne M, Hopper SD (2020) Contrasting patterns of population divergence on young and old landscapes in Banksia seminuda (Proteaceae), with evidence for recognition of subspecies. Biological Journal of the Linnean Society 133, 449–463.
| Contrasting patterns of population divergence on young and old landscapes in Banksia seminuda (Proteaceae), with evidence for recognition of subspecies.Crossref | GoogleScholarGoogle Scholar |
Sansaloni CP, Petroli CD, Carling J, Hudson CJ, Steane DA, Myburg AA, Grattapaglia D, Vaillancourt RE, Kilian A (2010) A high-density Diversity Arrays Technology (DArT) microarray for genome-wide genotyping in Eucalyptus. Plant Methods 6, 16
| A high-density Diversity Arrays Technology (DArT) microarray for genome-wide genotyping in Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH (2010) Integrative taxonomy: a multisource approach to exploring biodiversity. Annual Review of Entomology 55, 421–438.
| Integrative taxonomy: a multisource approach to exploring biodiversity.Crossref | GoogleScholarGoogle Scholar |
Scott J, Auld TD (2020) Conservation assessment of Prostanthera gilesii Althofer ex B.J.Conn & T.C.Wilson (family Lamiaceae). (NSW Department of Planning, Industry and Environment) Available at http://www.environment.gov.au/biodiversity/threatened/species/pubs/88743-conservation-advice-13112021.pdf
Searle SD, Bell JC, Moran GF (2000) Genetic diversity in natural populations of Acacia mearnsii. Australian Journal of Botany 48, 279–286.
| Genetic diversity in natural populations of Acacia mearnsii.Crossref | GoogleScholarGoogle Scholar |
Selwood KE, Zimmer HC (2020) Refuges for biodiversity conservation: a review of the evidence. Biological Conservation 245, 108502
| Refuges for biodiversity conservation: a review of the evidence.Crossref | GoogleScholarGoogle Scholar |
Silveira FAO, Fiedler PL, Hopper SD (2021) OCBIL theory: a new science for old ecosystems. Biological Journal of the Linnean Society 133, 251–265.
| OCBIL theory: a new science for old ecosystems.Crossref | GoogleScholarGoogle Scholar |
Slatyer RA, Nash MA, Miller AD, Endo Y, Umbers KD, Hoffmann AA (2014) Strong genetic structure corresponds to small-scale geographic breaks in the Australian alpine grasshopper Kosciuscola tristis. BMC Evolutionary Biology 14, 204
| Strong genetic structure corresponds to small-scale geographic breaks in the Australian alpine grasshopper Kosciuscola tristis.Crossref | GoogleScholarGoogle Scholar |
Smith JP, Heard TA, Beekman M, Gloag R (2017) Flight range of the Australian stingless bee Tetragonula carbonaria (Hymenoptera: Apidae). Austral Entomology 56, 50–53.
| Flight range of the Australian stingless bee Tetragonula carbonaria (Hymenoptera: Apidae).Crossref | GoogleScholarGoogle Scholar |
Sokal RR (1986) Phenetic Taxonomy: Theory and Methods. Annual Review of Ecology and Systematics 17, 423–442.
| Phenetic Taxonomy: Theory and Methods.Crossref | GoogleScholarGoogle Scholar |
Soltis DE, Gitzendanner MA, Stull G, Chester M, Chanderbali A, Chamala S, Jordon-Thaden I, Soltis PS, Schnable PS, Barbazuk WB (2013) The potential of genomics in plant systematics. Taxon 62, 886–898.
| The potential of genomics in plant systematics.Crossref | GoogleScholarGoogle Scholar |
Steane DA, Nicolle D, Sansaloni CP, Petroli CD, Carling J, Kilian A, Myburg AA, Grattapaglia D, Vaillancourt RE (2011) Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Molecular Phylogenetics and Evolution 59, 206–224.
| Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping.Crossref | GoogleScholarGoogle Scholar |
Sumner J, Haines ML, Lawrence P, Lawrence J, Clemann N (2021) Phylogenetic placement of a recently discovered population of the threatened alpine she-oak skink Cyclodomorphus praealtus (Squamata: Scincidae) in Victoria. Memoirs of Museum Victoria 153–157.
| Phylogenetic placement of a recently discovered population of the threatened alpine she-oak skink Cyclodomorphus praealtus (Squamata: Scincidae) in Victoria.Crossref | GoogleScholarGoogle Scholar |
Swofford D (2002) ‘PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods).’ (Sinauer Associates: Sunderland, MA, USA)
Symula R, Keogh JS, Cannatella DC (2008) Ancient phylogeographic divergence in southeastern Australia among populations of the widespread common froglet, Crinia signifera. Molecular Phylogenetics and Evolution 47, 569–580.
| Ancient phylogeographic divergence in southeastern Australia among populations of the widespread common froglet, Crinia signifera.Crossref | GoogleScholarGoogle Scholar |
Umbers KDL, Slatyer RA, Tatarnic NJ, Muschett GR, Wang S, Song H (2022) Phylogenetics of the skyhoppers (Kosciuscola) of the Australian Alps: evolutionary and conservation implications. Pacific Conservation Biology 28, 261–276.
| Phylogenetics of the skyhoppers (Kosciuscola) of the Australian Alps: evolutionary and conservation implications.Crossref | GoogleScholarGoogle Scholar |
van der Merwe MM, Yap J-YS, Wilson PD, Murphy HT, Ford A (2021) All populations matter: conservation genomics of Australia’s iconic purple wattle, Acacia purpureopetala. Diversity 13, 139
| All populations matter: conservation genomics of Australia’s iconic purple wattle, Acacia purpureopetala.Crossref | GoogleScholarGoogle Scholar |
von Mueller FJH (1858) Prostanthera phylicifolia. In ‘Fragmenta Phytographiae Australiae. Vol. 1’. p. 19. (Auctoritate Gubern: Melbourne, Vic., Australia)
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370.
| Estimating F-statistics for the analysis of population structure.Crossref | GoogleScholarGoogle Scholar |
Williams ML, Drinnan AN, Walsh NG (2006) Variation within Prostanthera spinosa (Lamiaceae): evidence from morphological and molecular studies. Australian Systematic Botany 19, 467–477.
| Variation within Prostanthera spinosa (Lamiaceae): evidence from morphological and molecular studies.Crossref | GoogleScholarGoogle Scholar |
Wilson TC, Conn BJ (2015) Two new species of Prostanthera (Lamiaceae) from south-eastern Queensland. Telopea 18, 255–263.
| Two new species of Prostanthera (Lamiaceae) from south-eastern Queensland.Crossref | GoogleScholarGoogle Scholar |
Wilson TC, Conn BJ, Henwood MJ (2012) Molecular phylogeny and systematics of Prostanthera (Lamiaceae). Australian Systematic Botany 25, 341–352.
| Molecular phylogeny and systematics of Prostanthera (Lamiaceae).Crossref | GoogleScholarGoogle Scholar |
Wilson TC, Conn BJ, Henwood MJ (2017) Great expectations: correlations between pollinator assemblages and floral characters in Lamiaceae. International Journal of Plant Sciences 178, 170–187.
| Great expectations: correlations between pollinator assemblages and floral characters in Lamiaceae.Crossref | GoogleScholarGoogle Scholar |
Wilson TC, Carmen P, Hook C (2019) Recircumscription of Prostanthera denticulata R.Br. (Lamiaceae, Westringieae) and the new species P. crocodyloides T.C.Wilson. Telopea 22, 75–87.
| Recircumscription of Prostanthera denticulata R.Br. (Lamiaceae, Westringieae) and the new species P. crocodyloides T.C.Wilson.Crossref | GoogleScholarGoogle Scholar |
Wilson TC, Rossetto M, Bain D, Yap J-YS, Wilson PD, Stimpson ML, Weston PH, Croft L (2022) A turn in species conservation for hairpin banksias: demonstration of oversplitting leads to better management of diversity. American Journal of Botany 109, 1652–1671.
| A turn in species conservation for hairpin banksias: demonstration of oversplitting leads to better management of diversity.Crossref | GoogleScholarGoogle Scholar |
Worth JRP, Marthick JR, Jordan GJ, Vaillancourt RE (2011) Low but structured chloroplast diversity in Atherosperma moschatum (Atherospermataceae) suggests bottlenecks in response to the Pleistocene glacials. Annals of Botany 108, 1247–1256.
| Low but structured chloroplast diversity in Atherosperma moschatum (Atherospermataceae) suggests bottlenecks in response to the Pleistocene glacials.Crossref | GoogleScholarGoogle Scholar |
Xu S, Li L, Luo X, Chen M, Tang W, Zhan L, Dai Z, Lam TT, Guan Y, Yu G (2022) ggtree: a serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 1, e56
| ggtree: a serialized data object for visualization of a phylogenetic tree and annotation data.Crossref | GoogleScholarGoogle Scholar |
Yap J-YS, Wilson TC, Rossetto M (2020) Conservation genomics of Prostanthera densa and P. marifolia: species status, management and translocation advice. November 2020 final report. (Research Centre for Ecosystem Resilience, Royal Botanic Garden: Sydney, NSW, Australia) Available at https://recer.org.au/wp-content/uploads/2022/09/Prostanthera-densa-and-P-marifolia-ReCER-Conservation-genomic-report.pdf
Yu G, Smith DK, Zhu H, Guan Y, Lam TT (2017) ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8, 28–36.
| ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data.Crossref | GoogleScholarGoogle Scholar |