Evolution of viviparity in mammals: what genomic imprinting tells us about mammalian placental evolution
Tomoko Kaneko-Ishino A C and Fumitoshi Ishino B C
B C
A School of Health Sciences, Tokai University, 143 Shimokasuya, Isehara, Kanagawa 259-1193, Japan.
B Department of Epigenetics, Medical Research Institute, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8510, Japan.
C Corresponding authors. Emails: fishino.epgn@mri.tmd.ac.jp; tkanekoi@is.icc.u-tokai.ac.jp
Reproduction, Fertility and Development 31(7) 1219-1227 https://doi.org/10.1071/RD18127
Submitted: 5 April 2018 Accepted: 1 October 2018 Published: 10 January 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Genomic imprinting is an epigenetic mechanism of regulating parent-of-origin-specific monoallelic expression of imprinted genes in viviparous therian mammals such as eutherians and marsupials. In this review we discuss several issues concerning the relationship between mammalian viviparity and genomic imprinting, as well as the domestication of essential placental genes: why has the genomic imprinting mechanism been so widely conserved despite the evident developmental disadvantages originating from monoallelic expression? How have genomic imprinted regions been established in the course of mammalian evolution? What drove the evolution of mammalian viviparity and how have genomic imprinting and domesticated genes contributed to this process? In considering the regulatory mechanism of imprinted genes, reciprocal expression of paternally and maternally expressed genes (PEGs and MEGs respectively) and the presence of several essential imprinted genes for placental formation and maintenance, it is likely that complementary, thereby monoallelic, expression of PEGs and MEGs is an evolutionary trade-off for survival. The innovation in novel imprinted regions was associated with the emergence of imprinting control regions, suggesting that genomic imprinting arose as a genome defence mechanism against the insertion of exogenous DNA. Mammalian viviparity emerged in the period when the atmospheric oxygen concentration was the lowest (~12%) during the last 550 million years (the Phanerozoic eon), implying this low oxygen concentration was a key factor in promoting mammalian viviparity as a response to a major evolutionary pressure. Because genomic imprinting and gene domestication from retrotransposons or retroviruses are effective measures of changing genomic function in therian mammals, they are likely to play critical roles in the emergence of viviparity for longer gestation periods.
Additional keywords: complementation hypothesis, domesticated genes, evolutionary trade-off, genome defence mechanism, Peg10, Peg11/Rtl1, reciprocal monoallelic expression, retrotransposons.
Why has genomic imprinting been widely conserved in viviparous mammals?
This question may be put alternatively as ‘What is the merit of genomic imprinting in the therian mammals that overwhelms its developmental disadvantages originating from monoallelic expression of a series of essential imprinted genes?’ All the human genomic imprinting diseases, such as Silver–Russell, Beckwith–Wiedemann, Prader–Willi, Angelman, Albright, Kagami–Ogata and Temple syndromes (Kalish et al. 2014), as well as their related developmental, growth and behavioural abnormalities observed in mice, are attributable to the monoallelic expression of imprinted genes. This is because (partial) uniparental chromosome duplications induce a lack and/or overexpression of relevant imprinted genes (Cattanach and Beechey 1990; Blake et al. 2010). Several advantages conferred by genomic imprinting have been proposed since its initial discovery. For example, the prevention of parthenogenetic development may be advantageous for mammalian reproduction because females can avoid childbearing in seasons unfit for child rearing (Solter 1988). Another advantage is the prevention of the development of trophoblast cells, placental cells with an invasive nature, to avoid developing malignant ovarian teratocarcinomas that arise during parthenogenesis (Varmuza and Mann 1994). However, it is difficult to test such hypotheses by purely experimental procedures. To address these types of issues, it is reasonable to expect that it will be necessary to at least advance our knowledge of the regulatory mechanism underlying genomic imprinting, the evolutionary relationship between genomic imprinting and viviparity, the environmental pressure that drove their evolution and the genetic factors that enabled these processes. Knowledge accumulated so far warrants further discussion.
In the two versions of ‘the conflict hypothesis’, the weak version has come to be widely accepted as the concept of ‘parental conflict’ (Moore and Haig 1991). This version states that in viviparous organisms, such as therian mammals, under polygamous conditions in which females are able to accept more than one male, the genes from the paternal alleles tend to promote embryo growth, whereas genes from the maternal alleles tend to inhibit embryo growth as a consequence of the genetic conflict between the paternal and maternal alleles. A substantial number of paternally and maternally expressed genes (PEGs and MEGs respectively) fit well with this prediction, such as the paternally expressed insulin-like growth factor 2 (IGF2) and maternally expressed insulin-like growth factor 2 receptor (IGF2R) genes (Killian et al. 2000; Reik and Walter 2001; Wilkins and Haig 2003; Hore et al. 2007; Renfree et al. 2009). The former encodes the embryonic growth factor IGF2, whereas IGF2R protein binds IGF2 and degrades it by transporting it to the lysosome. Conversely, the so-called strong version of the conflict hypothesis predicts that under the continuous pressure exerted by genetic conflict, the genomic imprinting mechanism inevitably originates in viviparous organisms during the course of evolution (Wilkins and Haig 2003). Through a process known as evolutionary equilibrium, a locus that was initially expressed in both alleles gradually comes to be expressed from a single allele, with the other allele made silent by means of natural selection. This process involves the increased expression of one allele and the decreased expression of the other. For example, a growth factor gene initially showing biallelic expression is gradually changed to paternal allele-specific expression over a long period, when fitness is determined by the level of the growth factor and natural selection favours higher production when an allele is paternally derived than when it is maternally derived. It is known that genomic imprinting is also observed in plants, such as angiosperms, providing strong support for this hypothesis (Haig and Westoby 1991). However, the emergence of genomic imprinting regions in mammals is likely to arise as a genome defence mechanism against the insertion of exogenous DNA, suggesting that target chromosomal regions and/or genes were at first randomly mutated. Some were then selected as imprinted regions and/or imprinted genes as a result of developmental advantages, which has also been predicted by the weak version of the conflict hypothesis.
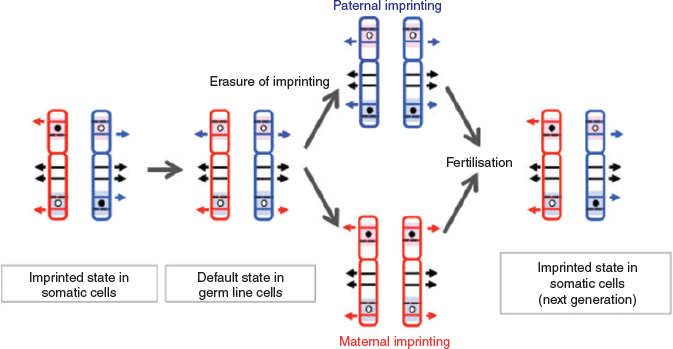
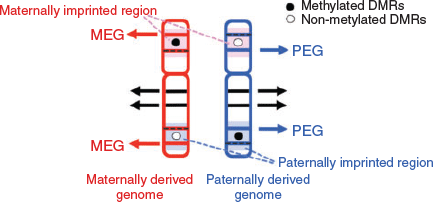
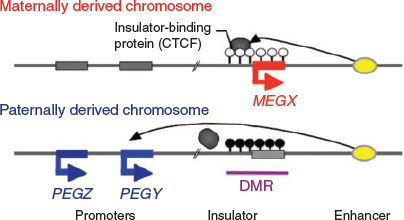
In considering the regulatory mechanism of imprinted gene expression, we have proposed ‘the complementary hypothesis’ (Kaneko-Ishino et al. 2003, 2006; Kaneko-Ishino and Ishino 2015), in which the monoallelic expression mechanism emerged as an evolutionary trade-off for survival. Usually, each imprinted region comprises both PEGs and MEGs and their reciprocal expression is regulated by the DNA methylation status of each differentially methylated region (DMR; also known as an imprinting control region (ICR); Fig. 1). The imprinted regions are divided into two groups, namely paternally and maternally imprinted regions, with DNA methylation in the paternal and maternal alleles respectively. The DMR sequences seem to function as cis-regulatory units, such as acting as an insulator (IGF2–H19, imprinted maternal expressed transcript (H19) region; Bell and Felsenfeld 2000; Hark et al. 2000) or transcriptional start site of antisense RNA (antisense of IGF2R non-protein coding RNA (AIRN) in the IGF2R region; Sleutels et al. 2002; Latos et al. 2012; Fig. 2). This is the mechanism by which the methylated DMR sequence not only represses certain imprinted genes, but also induces expression of other imprinted genes over a substantially wide imprinted region, sometimes up to 500 Mb. In the maternally imprinted regions, DMR DNA methylation represses PEGs but induces MEGs. Conversely, DMR DNA methylation in paternally imprinted regions represses MEGs while inducing PEGs (Figs 1, 2). Thus, it is clear that PEGs and MEGs cannot be expressed from the individual parental chromosomes simultaneously; that is, two chromosomes of different DNA methylation status (i.e. two different epigenotypes) are necessary for the simultaneous expression of PEGs and MEGs in somatic cells.
|
|
Using somatic cloning techniques, cloned embryos generated from the nuclei of primordial germ cells on embryonic Day 12 represent the default state of genomic imprinting; that is, both parental alleles lose all DMR DNA methylation and thus have the same status. Importantly, they exhibit early embryonic lethality like parthenogenetic and androgenetic embryos, supporting the concept that two different parental epigenotypes are essential for development (Lee et al. 2002). Under this condition, only PEGs are expressed from the maternally imprinted regions and only MEGs are expressed from the paternally imprinted regions, so they lack expression of several essential imprinted genes (Fig. 3).
|
This provides a notion of perceptual shift, because most hypotheses on genomic imprinting presuppose that PEGs and MEGs exhibit monoallelic expression under the control of genomic imprinting and are expressed from both parental alleles equally (biallelic expression) without any particular regulation. As discussed later, a substantial number of PEGs and MEGs play essential roles in development, growth and behaviour, indicating that complementary monoallelic expression is essential for survival in the current adaptive form of therian mammals. Thus, it is likely that it arose as an evolutionary trade-off for survival against the disadvantageous monoallelic expression of several essential and/or important developmental genes.
How have genomic imprinting regions been established in mammals?
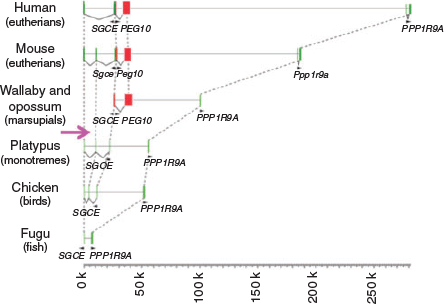
More than 15 imprinted regions exist in the genomes of humans and mice, and each region has at least one DMR. Unexpectedly, no sequence homology was identified among the DMRs in either the paternally or maternally imprinted regions. This has hampered understanding of the common features of the DMRs, particularly their regulatory mechanisms. An important clue to how imprinted regions emerged during evolution came from a series of comprehensive comparative studies of imprinted genes and/or imprinted regions between eutherians and marsupials. In marsupials, only six imprinted genes, namely IGF2, IGF2R, paternally expressed 1 (PEG1)/mesoderm specific transcript (MEST), paternally expressed 10 (PEG10), insulin (INS) and H19, have thus far been identified, whereas many other orthologues of eutherian imprinted genes are not imprinted (Renfree et al. 2009, 2012). Of these imprinted genes, the PEG10 and IGF2–H19 regions have conserved DMRs (PEG10 and H19 DMRs) in both eutherians and marsupials, demonstrating that at least these two regions existed in a common therian ancestor (Suzuki et al. 2007; Smits et al. 2008). These observations also demonstrate that those DMRs were present at the beginning of the genomic imprinting history, and that several imprinted genes and imprinted regions were considerably increased later in the eutherian lineage. In addition to their DMRs, the PEG10 and H19 genes first emerged in the therian genomes, because there are no corresponding DNA sequences present in platypus (monotremes) or other non-mammal vertebrates, such as the chicken (birds) and fugu (fish). The evidence further indicates that it was newly inserted exogenous DNA in these regions from which the PEG10 and H19 DMRs originated (Renfree et al. 2012; Kaneko-Ishino and Ishino 2015) (Figs 4, 5).
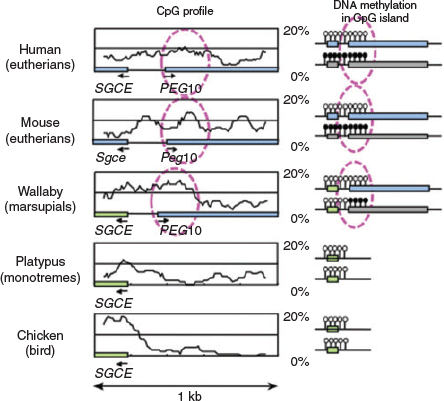
|
The same is true for most of the DMRs in the eutherian-specific imprinted regions (Suzuki et al. 2011; Renfree et al. 2012; Fig. 6), except for small nuclear ribonucleoprotein polypeptide N (SNRPN) and solute carrier family 38 member 4 (SLC38A4) DMRs, in which the DNA sequences pre-existed in a common mammalian and therian ancestor respectively. In both cases, eutherian-specific genomic imprinting regulation was associated with eutherian-specific chromosomal translocation that occurred in the neighbourhood of each DMR (Rapkins et al. 2006; Suzuki et al. 2011). Some of the imprinted regions, such as malignant T cell amplified sequence 2 (Mcts2), paternally expressed 13 (Peg13), Ras protein specific guanine nucleotide releasing factor 1 (Rasgrf1), zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 (Zrsrt)/U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1 (U2af1-rs1) and impact, RWD protein (Impact), emerged in a lineage-specific manner, in which the insertion of their unique DMR sequences was confirmed. Thus, one conclusion that could be drawn is that the emergence of genomic imprinting regions occurred in association with major chromosomal changes (i.e. the insertion of exogenous DNA and/or chromosome translocation). This implies that genomic imprinting arose as a genome defence mechanism against the disturbance of the local genetic network by cis-regulatory units, probably due to either the loss or addition of such unit(s). As mentioned above, the complementary monoallelic expression system of genomic imprinting may be a useful means to overcome these situations, which could otherwise result in developmental abnormalities and/or lethality. Thus, the number of imprinted regions may only represent a list of successful recovery of genomic imprinting. Many more insertion and/or chromosomal rearrangement events must have occurred at the level of the complete genome, but harmful loci were lost, whereas non-harmful loci have remained. It should be noted that several imprinted genes that reside in the introns of other genes, such as Mcts2, nucleosome assembly protein 1 like 5 (Nap1L5), inositol polyphosphate-5-phosphatase F variant 2 (Inpp5f_v2), U2af1-rs1 and neuronatin (Nnat), are duplicated genes that were inserted by cDNA retrotransposition and their promoter regions became their DMRs (Cowley and Oakey 2010).
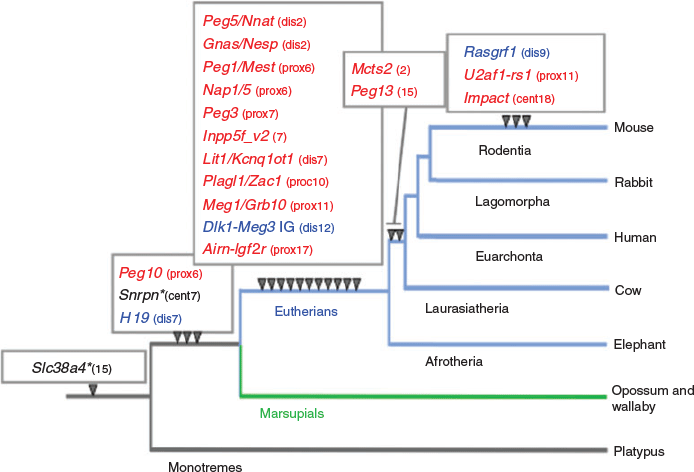
|
There are no conserved DMRs in the marsupial PEG1/MEST or IGF2R region (Suzuki et al. 2005; Weidman et al. 2006). It is of considerable interest to elucidate how these genes were imprinted, although the latter may be regulated by a different DMR than that in eutherians. One possibility is histone modification, which functions as an alternative regulatory mechanism of genomic imprinting. Both eutherians and marsupials have another epigenetic mechanism of X chromosome inactivation (XCI; Lyon 1963; and 1986). One of two female X chromosomes is inactivated by expression of X inactive specific transcript (XIST), a non-coding RNA specifically expressed from an inactivated X chromosome. At least in eutherians, H3K27 trimethylation, not DNA methylation, inactivates XIST expression from an active X chromosome, leading to monoallelic XIST expression, similar to that in genomic imprinting (Inoue et al. 2017a). Both histone modification and DNA methylation result in genomic imprinting (Li et al. 2008; Inoue et al. 2017b), but in the eutherians the control of genomic imprinting must have shifted to the latter, probably because a strict all-or-none-type of regulation was more advantageous. In this regard, acquisition of DNA methyltransferase 3 like (DNMT3L) in the therian lineage may be of critical importance (Yokomine et al. 2006) because it regulates DNA methylation in germ cells in cooperation with DNA methyltransferase 3A and B (DNMT3A and DNMT3B), thus establishing the DNA methylation status of genome imprinting (Bourc’his et al. 2001; Hata et al. 2002).
Relationship between genomic imprinting and viviparous reproduction
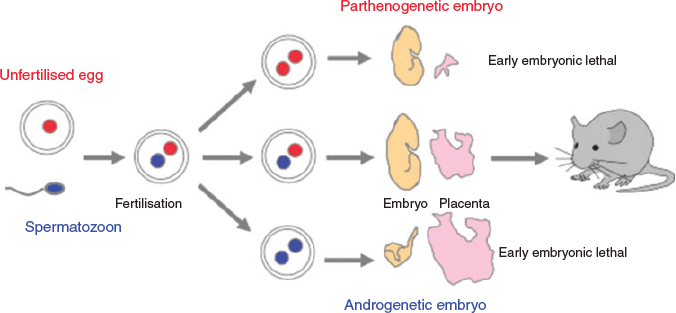
As mentioned earlier, among the vertebrates, genomic imprinting is unique to viviparous mammals. It was originally discovered in 1984 from experiments showing that parthenogenetic and androgenetic embryos with either two maternal or two paternal pronuclei exhibited early embryonic lethality, albeit with different morphological anomalies in the placenta (Surani et al. 1984; Mann and Lovell-Badge 1984; McGrath and Solter 1984; Fig. 7). The former died at an early embryonic stage due to severe placental defects, even though the embryos appeared normal. The latter also exhibited early embryonic lethality due to severe growth retardation along with overgrowth of the impaired placenta. This not only demonstrates the functional differences between the paternal and maternal genomes, but also suggests a profound relationship between genomic imprinting and placental formation and function.
Androgenetic death is attributable to at least two imprinted genes, paternally expressed Igf2 and maternally expressed achaete-scute family b HLH transcription factor 2 (Ascl2)/murine achaete-scute homolog 2 (Mash2), which are located in two closely neighbouring imprinted regions on the mouse distal chromosome 7 (Cattanach and Beechey 1990; Blake et al. 2010; Fig. 8). Ascl2/Mash2 encodes a placenta-specific transcription factor essential for the formation of placental spongiotrophoblast and labyrinth layers (Guillemot et al. 1994, 1995). A lack of Ascl2/Mash2 combined with overproduction of embryonic growth hormone Igf2 (Constância et al. 2002) explains the placental phenotype of the androgenetic embryos. Ascl2/Mash2 emerged as a duplicated gene of achaete-scute family b HLH transcription factor 1 (Ascl1)/murine achaete-scute homolog 1 (Mash1), encoding a brain-specific transcription factor. In chickens, both are expressed in the brain, whereas Ascl2/Mash2 exhibits placenta-specific expression, at least in humans and mice, suggesting that it was recruited for placenta formation in the course of eutherian evolution (Yokomine et al. 2005).
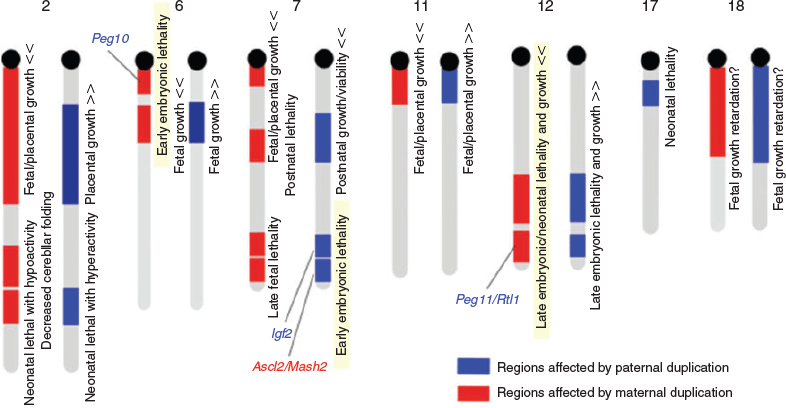
|
Mouse proximal chromosome 6 is an imprinted region, the maternal duplication of which causes early embryonic lethality. Peg10 in this region is the major gene responsible for the early embryonic lethal phenotype. Peg10-knockout (KO) embryos die due to severe placental defects, as for parthenogenetic embryos (Ono et al. 2006). The placentas of Peg10-KO embryos lack spongiotrophoblast and labyrinth layers, as seen in Ascl2/Mash2-KO mice. It should be noted that this relationship between Peg10 and Ascl2/Mash2 is an apparent exception to the ‘parental conflict’ model, because both the PEGs and MEGs exhibit very similar embryonic growth promotion. As described earlier, PEG10 is a therian-specific gene as well as one of the first imprinted genes to appear in a common therian ancestor. Importantly, PEG10 is a long terminal repeat (LTR) retrotransposon-derived gene encoding an LTR retrotransposon Gag- and Pol-like protein (Ono et al. 2001). Thus, this is a very good example of a therian-specific gene playing an essential role in the formation of the placenta, a therian-specific organ for viviparous reproduction.
Viviparous reproduction is widely observed in vertebrates and non-vertebrates, such as squamates, sharks and scorpions. However, a unique feature of the mammalian placenta is the inclusion of trophoblast cells (Roberts et al. 2016). Eutherians and marsupials developed different types of placentas (i.e. chorioallantoic and choriovitelline (yolk sac) placentas respectively) and both contain trophoblast cells in addition to the chorion, allantois or yolk sac membranes. It is highly probable that PEG10 plays an essential role in the differentiation of such trophoblast cells in the spongiotrophoblast and labyrinth layers. If this is the case, domestication of PEG10 was a critical event in the establishment of the current therian viviparous systems.
Another important question concerning therian viviparity is why eutherians and marsupials respectively adopted chorioallantoic and yolk sac placentas (Renfree 2010). Some eutherian species, such as dogs and cats, also use yolk sac placentas in the early stage of gestation, whereas some marsupial species, such as bandicoots, develop chorioallantoic placentas, which they use for a short period in the very last stage of gestation (Tyndale-Biscoe and Renfree 1987). This suggests that both groups had an equal chance to widely adopt the chorioallantoic placenta. PEG11/retrotransposon Gaglike 1 (RTL1), another paternally expressed imprinted gene, may key to addressing this question (Charlier et al. 2001). Mouse Peg11/Rtl1 located on the distal chromosome 12 is a major gene responsible for late embryo or neonatal lethality and is associated with late embryo growth retardation upon maternal duplication of this region (Sekita et al. 2008). Peg11/Rtl1 plays an essential role in ‘the feto–maternal interface’ in the placenta; that is, in fetal capillary endothelial cells, where nutrients and oxygen are exchanged between the fetal and maternal blood. The fetal capillary endothelial cells are surrounded by three layers of trophoblast cells, two syncytiotrophoblast layers and one cytotrophoblast layer. The trophoblast cells could harm the maternal tissues because of their invasive nature. In the Peg11/Rtl1-KO placenta, the fetal capillary endothelial cells were found to be severally damaged by trophoblast cells, whereas the trophoblast themselves were severally damaged when Peg11/Rtl1 was overproduced (Sekita et al. 2008). Therefore, Peg11/Rtl1 is essential for the maintenance of the eutherian-type of chorioallantoic placenta for the entire gestation period. Importantly, PEG11/RTL1 is another LTR retrotransposon-derived gene specific to eutherians that is absent from marsupials (Charlier et al. 2001; Edwards et al. 2008). This explains why chorioallantoic placentas were not widely adopted in marsupial species, at least partly because a short-lived chorioallantoic organ may not have enough advantage to override the adoption of the yolk sac placenta. Thus, elucidation of the biochemical function of PEG11/RTL1 is important to understanding the evolutionary history of how the viviparous systems diverged in eutherians and marsupials.
What drove the evolution of viviparity in mammals?
As mentioned above, the emergence of therian mammals 168 million years before the present (Ma) coincided with the appearance of genomic imprinting and domestication of one of the essential placental genes, PEG10. Both events may have had a considerable effect on genomic function, especially in association with the acquisition of viviparity. The divergence of the eutherians and marsupials 148 Ma coincided with the emergence of a series of new imprinted regions and the domestication of another essential placental gene, PEG11/RTL1. Both events may have affected a wide range of eutherian and marsupial genomic functions relating to the diversification of the viviparous reproduction systems. Thus, genomic imprinting and gene domestication may have played critical roles in the evolution and diversification of the viviparous reproduction systems as effective adaptive measures. However, in the first place, what kind of environmental factor(s) drove the evolution of mammalian viviparity? That is, what was the major evolutionary pressure that forced the therian mammals to innovate a new style of reproduction system?
One possibility is the low atmospheric oxygen concentration (12–13%) approximately 190–150 Ma in the Phanerozoic eon (from 550 Ma to the present; Berner et al. 2007), because oxygen concentration is a major factor affecting development and growth in a wide range of organisms, including plants and animals. However, it is also possible that this was a coincidence. Two consecutive mass extinction events occurred at the Permian–Triassic and Triassic–Jurassic boundaries 250 and 200 Ma respectively as a result of low atmospheric oxygen, most of the therapsids, except for the ancestors of mammals, became extinct. Dinosaurs, ancestors of birds, adapted to these conditions using an air sac system with high oxygen exchange efficiency and dominated the world for a long time. It is likely that an efficient oxygen supply to the embryo via a placental blood circulating system along with an effective nutrient supply would provide a developmental advantage.
Conclusion: evolutionary trade-off in mammals
Viviparous reproduction exerts an inordinate burden on females. Even today, many women die due to problems related to pregnancy and childbirth. Humans and rodents have the most invasive type of haemochorial placentas. Interestingly, recent phylogenetic analysis suggests that the haemochorial or endotheliochorial placenta that has a discoid shape with labyrinths, but not villi, represents the ancestral type of the eutherian placenta, whereas a non-invasive type of epithetiochorial placenta is an evolution-derived phenotype for avoiding immunological problems between the mother and hemiallogeneic fetus (Wildman et al. 2006; Roberts et al. 2016). It is likely that the adoption of viviparity using the invasive type of placenta was a major challenge for the common eutherian ancestor in the course of adapting to low atmospheric oxygen but must have the invasive type of placenta conferred considerable advantages to override the dangers arising from viviparous reproduction in females. It could be concluded that the acquisition of viviparity was an evolutionary trade-off in eutherian mammals.
Retrospectively, the long gestation period resulting from the adoption of a chorioallantoic placenta allowed extreme brain development in eutherians, whereas marsupials achieved a comparable brain size through an extended lactation period (Ashwell 2008; Weisbecker and Goswami 2010). It has become apparent that several imprinted genes (Keverne 2013, 2014) and several LTR retrotransposon-derived genes (Kaneko-Ishino and Ishino 2012, 2015) play important roles in a variety of brain functions. Thus, genomic imprinting and domestication of novel genes have proven to be effective measures for the innovation of therian- and eutherian-specific traits, including viviparous reproduction and sophisticated brain systems. Evolutionary events associated with genomic imprinted regions and domesticated genes appear to have emerged by chance, and were subsequently fixed in mammalian genomes by natural selection, as Darwin predicted. In most cases, some sort of evolutionary trade-off must have been involved in the process of adapting to new environments. This helps explain why genomic imprinting at first looked so complicated from an evolutionary viewpoint.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors thank all their collaborators and laboratory members, especially Azim Surani, Marilyn Renfree, Atsuo Ogura, Takashi Kohda, Jiyoung Lee, Ryu-ichi Ono, Yoichi Sekita, Shunsuke Suzuki, Hirosuke Shiura and Moe Kitazawa.
References
Ashwell, K. W. (2008). Encephalization of Australian and New Guinean marsupials. Brain Behav. Evol. 71, 181–199.| Encephalization of Australian and New Guinean marsupials.Crossref | GoogleScholarGoogle Scholar | 18230970PubMed |
Bell, A. C., and Felsenfeld, G. (2000). Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485.
| Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene.Crossref | GoogleScholarGoogle Scholar | 10839546PubMed |
Berner, R. A., Vandenbrooks, J. M., and Ward, P. D. (2007). Evolution. Oxygen and evolution. Science 316, 557–558.
| Evolution. Oxygen and evolution.Crossref | GoogleScholarGoogle Scholar | 17463279PubMed |
Blake, A., Pickford, K., Greenaway, S., Thomas, S., Pickard, A., Williamson, C. M., Adams, N. C., Walling, A., Beck, T., Fray, M., Peters, J., Weaver, T., Brown, S. D., Hancock, J. M., and Mallon, A. M. (2010). MouseBook: an integrated portal of mouse resources. Nucleic Acids Res. 38, D593–D599.
| MouseBook: an integrated portal of mouse resources.Crossref | GoogleScholarGoogle Scholar | 19854936PubMed |
Bourc’his, D., Xy, G. L., Lin, C. S., Bollman, B., and Bester, T. H. (2001). Dmnt3L and the establishment of maternal genomic imprinting. Science 294, 2536–2539.
| Dmnt3L and the establishment of maternal genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 11719692PubMed |
Cattanach, B. M., and Beechey, C. V. (1990). Autosomal and X-chromosome imprinting. Development 108, 63–72.
Charlier, C., Segers, K., Wagenaar, D., Karim, L., Berghmans, S., Jaillon, O., Shay, T., Weissenbach, J., Cockett, N., Gyapay, G., and Georges, M. (2001). Human–ovine comparative sequencing of a 250-kb imprinted domain encompassing the Callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11, 850–862.
| Human–ovine comparative sequencing of a 250-kb imprinted domain encompassing the Callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8.Crossref | GoogleScholarGoogle Scholar | 11337479PubMed |
Constância, M., Hemberger, M., Hughes, J., Dean, W., Ferguson-Smith, A., Fundele, R., Stewart, F., Kelsey, G., Fowden, A., Sibley, C., and Reik, W. (2002). Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417, 945–948.
| Placental-specific IGF-II is a major modulator of placental and fetal growth.Crossref | GoogleScholarGoogle Scholar | 12087403PubMed |
Cowley, M., and Oakey, R. J. (2010). Retrotransposition and genomic imprinting. Brief Funct. Genomics 9, 340–346.
| Retrotransposition and genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 20591835PubMed |
Edwards, C. A., Mungall, A. J., Matthews, L., Ryder, E., Gray, D. J., Pask, A. J., Shaw, G., Graves, J. A., Rogers, J., SAVOIR consortium Dunham, I., Renfree, M. B., and Ferguson-Smith, A. C. (2008). The evolution of the DLK1–DIO3 imprinted domain in mammals. PLoS Biol. 6, e135.
| The evolution of the DLK1–DIO3 imprinted domain in mammals.Crossref | GoogleScholarGoogle Scholar | 18532878PubMed |
Guillemot, F., Nagy, A., Auerbach, A., Rossant, J., and Joyner, A. L. (1994). Essential role of Mash-2 in extraembryonic development. Nature 371, 333–336.
| Essential role of Mash-2 in extraembryonic development.Crossref | GoogleScholarGoogle Scholar | 8090202PubMed |
Guillemot, F., Caspary, T., Tilghman, S. M., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Anderson, D. J., Joyner, A. L., Rossant, J., and Nagy, A. (1995). Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9, 235–242.
| Genomic imprinting of Mash2, a mouse gene required for trophoblast development.Crossref | GoogleScholarGoogle Scholar | 7773285PubMed |
Haig, D., and Westoby, M. (1991). Genomic imprinting in endosperm: its effects on seed development in crosses between species and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333, 1–13.
| Genomic imprinting in endosperm: its effects on seed development in crosses between species and between different ploidies of the same species, and its implications for the evolution of apomixis.Crossref | GoogleScholarGoogle Scholar |
Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M., and Tilghman, S. M. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489.
| CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus.Crossref | GoogleScholarGoogle Scholar | 10839547PubMed |
Hata, K., Okano, M., Lei, H., and Li, E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltranferase to establish maternal imprints in mice. Development 129, 1983–1993.
| 11934864PubMed |
Hore, T. A., Rapkins, R. W., and Graves, J. A. (2007). Construction and evolution of imprinted loci in mammals. Trends Genet. 23, 440–448.
| Construction and evolution of imprinted loci in mammals.Crossref | GoogleScholarGoogle Scholar | 17683825PubMed |
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017a). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424.
| Maternal H3K27me3 controls DNA methylation-independent imprinting.Crossref | GoogleScholarGoogle Scholar | 28723896PubMed |
Inoue, A., Jiang, L., Lu, F., and Zhang, Y. (2017b). Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 31, 1927–1932.
| Genomic imprinting of Xist by maternal H3K27me3.Crossref | GoogleScholarGoogle Scholar | 29089420PubMed |
Kalish, J. M., Jiang, C., and Bartolomei, M. S. (2014). Epigenetics and imprinting in human diseases. Int. J. Dev. Biol. 58, 291–298.
| Epigenetics and imprinting in human diseases.Crossref | GoogleScholarGoogle Scholar | 25023695PubMed |
Kaneko-Ishino, T., and Ishino, F. (2012). The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 3, 262.
| The role of genes domesticated from LTR retrotransposons and retroviruses in mammals.Crossref | GoogleScholarGoogle Scholar | 22866050PubMed |
Kaneko-Ishino, T., and Ishino, F. (2015). Mammalian-specific genomic functions: newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 91, 511–538.
| Mammalian-specific genomic functions: newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals.Crossref | GoogleScholarGoogle Scholar | 26666304PubMed |
Kaneko-Ishino, T., Kohda, T., and Ishino, F. (2003). The regulation and biological significance of genomic imprinting in mammals. J. Biochem. 133, 699–711.
| The regulation and biological significance of genomic imprinting in mammals.Crossref | GoogleScholarGoogle Scholar | 12869525PubMed |
Kaneko-Ishino, T., Kohda, T., Ono, R., and Ishino, F. (2006). Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet. Genome Res. 113, 24–30.
| Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development.Crossref | GoogleScholarGoogle Scholar | 16575159PubMed |
Keverne, E. B. (2013). Importance of the matriline for genomic imprinting, brain development and behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110327.
| Importance of the matriline for genomic imprinting, brain development and behaviour.Crossref | GoogleScholarGoogle Scholar | 23166391PubMed |
Keverne, E. B. (2014). Mammalian viviparity: a complex niche in the evolution of genomic imprinting. Heredity 113, 138–144.
| Mammalian viviparity: a complex niche in the evolution of genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 24569636PubMed |
Killian, J. K., Byrd, J. C., Jirtle, J. V., Munday, B. L., Stoskopf, M. K., MacDonald, R. G., and Jirtle, R. L. (2000). M6P/IGF2R imprinting evolution in mammals. Mol. Cell 5, 707–716.
| M6P/IGF2R imprinting evolution in mammals.Crossref | GoogleScholarGoogle Scholar | 10882106PubMed |
Latos, P. A., Pauler, F. M., Koerner, M. V., Şenergin, H. B., Hudson, Q. J., Stocsits, R. R., Allhoff, W., Stricker, S. H., Klement, R. M., Warczok, K. E., Aumayr, K., Pasierbek, P., and Barlow, D. P. (2012). Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472.
| Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing.Crossref | GoogleScholarGoogle Scholar | 23239737PubMed |
Lee, J., Inoue, K., Ono, R., Ogonuki, N., Kohda, T., Kaneko-Ishino, T., Ogura, A., and Ishino, F. (2002). Erasing genomic imprinting memory in mouse clone embryos produced from Day-11.5 primordial germ cells. Development 129, 1807–1817.
| 11934847PubMed |
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., and Ferguson-Smith, A. C. (2008). A maternal–zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557.
| A maternal–zygotic effect gene, Zfp57, maintains both maternal and paternal imprints.Crossref | GoogleScholarGoogle Scholar | 18854139PubMed |
Lyon, M. F. (1963). Lyonization of the X chromosome. Lancet 282, 1120–1121.
| Lyonization of the X chromosome.Crossref | GoogleScholarGoogle Scholar |
Lyon, M. F. (1986). X chromosomes and dosage compensation. Nature 320, 313.
| X chromosomes and dosage compensation.Crossref | GoogleScholarGoogle Scholar | 3960115PubMed |
Mann, J. R., and Lovell-Badge, R. H. (1984). Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature 310, 66–67.
| Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm.Crossref | GoogleScholarGoogle Scholar | 6738704PubMed |
McGrath, J., and Solter, D. (1984). Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183.
| Completion of mouse embryogenesis requires both the maternal and paternal genomes.Crossref | GoogleScholarGoogle Scholar | 6722870PubMed |
Moore, T., and Haig, D. (1991). Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7, 45–49.
| Genomic imprinting in mammalian development: a parental tug-of-war.Crossref | GoogleScholarGoogle Scholar | 2035190PubMed |
Ono, R., Kobayashi, S., Wagatsuma, H., Aisaka, K., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2001). A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237.
| A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21.Crossref | GoogleScholarGoogle Scholar | 11318613PubMed |
Ono, R., Nakamura, K., Inoue, K., Naruse, M., Usami, T., Wakisaka-Saito, N., Hino, T., Suzuki-Migishima, R., Ogonuki, N., Miki, H., Kohda, T., Ogura, A., Yokoyama, M., Kaneko-Ishino, T., and Ishino, F. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106.
| Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality.Crossref | GoogleScholarGoogle Scholar | 16341224PubMed |
Rapkins, R. W., Hore, T., Smithwick, M., Ager, E., Pask, A. J., Renfree, M. B., Kohn, M., Hameister, H., Nicholls, R. D., Deakin, J. E., and Graves, J. A. (2006). Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet. 2, e182.
| Recent assembly of an imprinted domain from non-imprinted components.Crossref | GoogleScholarGoogle Scholar | 17069464PubMed |
Reik, W., and Walter, J. (2001). Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32.
| Genomic imprinting: parental influence on the genome.Crossref | GoogleScholarGoogle Scholar | 11253064PubMed |
Renfree, M. B. (2010). Review: marsupials: placental mammals with a difference. Placenta 31, S21–S26.
| Review: marsupials: placental mammals with a difference.Crossref | GoogleScholarGoogle Scholar | 20079531PubMed |
Renfree, M. B., Hore, T. A., Shaw, G., Graves, J. A., and Pask, A. J. (2009). Evolution of genomic imprinting: insights from marsupials and monotremes. Annu. Rev. Genomics Hum. Genet. 10, 241–262.
| Evolution of genomic imprinting: insights from marsupials and monotremes.Crossref | GoogleScholarGoogle Scholar | 19630559PubMed |
Renfree, M. B., Suzuki, S., and Kaneko-Ishino, T. (2012). The origin and evolution of genomic imprinting in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120151.
Roberts, R. M., Green, J. A., and Schulz, L. C. (2016). The evolution of the placenta. Reproduction 152, R179–R189.
| The evolution of the placenta.Crossref | GoogleScholarGoogle Scholar | 27486265PubMed |
Sekita, Y., Wagatsuma, H., Nakamura, K., Ono, R., Kagami, M., Wakisaka, N., Hino, T., Suzuki-Migishima, R., Kohda, T., Ogura, A., Ogata, T., Yokoyama, M., Kaneko-Ishino, T., and Ishino, F. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto–maternal interface of mouse placenta. Nat. Genet. 40, 243–248.
| Role of retrotransposon-derived imprinted gene, Rtl1, in the feto–maternal interface of mouse placenta.Crossref | GoogleScholarGoogle Scholar | 18176565PubMed |
Sleutels, F., Zwart, R., and Barlow, D. P. (2002). The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813.
| The non-coding Air RNA is required for silencing autosomal imprinted genes.Crossref | GoogleScholarGoogle Scholar | 11845212PubMed |
Smits, G., Mungall, A. J., Griffiths-Jones, S., Smith, P., Beury, D., Matthews, L., Rogers, J., Pask, A. J., Shaw, G., VandeBerg, J. L., McCarrey, J. R., SAVOIR Consortium Renfree, M. B., Reik, W., and Dunham, I. (2008). Conservation of the H19 noncoding RNA and H19–IGF2 imprinting mechanism in therians. Nat. Genet. 40, 971–976.
| Conservation of the H19 noncoding RNA and H19–IGF2 imprinting mechanism in therians.Crossref | GoogleScholarGoogle Scholar | 18587395PubMed |
Solter, D. (1988). Differential imprinting and expression of maternal and paternal genomes. Annu. Rev. Genet. 22, 127–146.
| Differential imprinting and expression of maternal and paternal genomes.Crossref | GoogleScholarGoogle Scholar | 3071246PubMed |
Surani, M. A., Barton, S. C., and Norris, M. L. (1984). Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550.
| Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis.Crossref | GoogleScholarGoogle Scholar | 6709062PubMed |
Suzuki, S., Renfree, M. B., Pask, A., Shaw, G., Kobayashi, S., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2005). Genomic imprinting of IGF2, P57KIP2 and PEG1/MEST in a marsupial, the tammar wallaby. Mech. Dev. 122, 213–222.
| Genomic imprinting of IGF2, P57KIP2 and PEG1/MEST in a marsupial, the tammar wallaby.Crossref | GoogleScholarGoogle Scholar | 15652708PubMed |
Suzuki, S., Ono, R., Narita, T., Pask, A. J., Shaw, G., Wang, C., Kohda, T., Alsop, A. E., Graves, M. J. A., Kohara, Y., Ishino, F., Renfree, M. B., and Kaneko-Ishino, T. (2007). Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3, e55.
| Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 17432937PubMed |
Suzuki, S., Shaw, G., Kaneko-Ishino, T., Ishino, F., and Renfree, M. B. (2011). The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands. Genome Biol. Evol. 3, 1276–1283.
| The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands.Crossref | GoogleScholarGoogle Scholar | 22016334PubMed |
Tyndale-Biscoe, H., and Renfree, M. (1987). Monographs on marsupial biology. In ‘Reproductive Physiology of Marsupials’. pp. 310–323. (Cambridge University Press: Cambridge.)
Varmuza, S., and Mann, M. (1994). Genomic imprinting – defusing the ovarian time bomb. Trends Genet. 10, 118–123.
| Genomic imprinting – defusing the ovarian time bomb.Crossref | GoogleScholarGoogle Scholar | 7848407PubMed |
Weidman, J. R., Dolinoy, D. C., Maloney, K. A., Cheng, J. F., and Jirtle, R. L. (2006). Imprinting of opossum Igf2r in the absence of differential methylation and air. Epigenetics 1, 50–55.
| Imprinting of opossum Igf2r in the absence of differential methylation and air.Crossref | GoogleScholarGoogle Scholar |
Weisbecker, V., and Goswami, A. (2010). Brain size, life history, and metabolism at the marsupial/placental dichotomy. Proc. Natl Acad. Sci. USA 107, 16216–16221.
| Brain size, life history, and metabolism at the marsupial/placental dichotomy.Crossref | GoogleScholarGoogle Scholar | 20823252PubMed |
Wildman, D. E., Chen, C., Erez, O., Grossman, L. I., Goodman, M., and Romero, R. (2006). Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl Acad. Sci. USA 103, 3203–3208.
| Evolution of the mammalian placenta revealed by phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar | 16492730PubMed |
Wilkins, J. F., and Haig, D. (2003). What good is genomic imprinting: the function of parent-specific gene expression. Nat. Rev. Genet. 4, 359–368.
| What good is genomic imprinting: the function of parent-specific gene expression.Crossref | GoogleScholarGoogle Scholar | 12728278PubMed |
Yokomine, T., Shirohzu, H., Purbowasito, W., Toyoda, A., Iwama, H., Ikeo, K., Hori, T., Mizuno, S., Tsudzuki, M., Matsuda, Y., Hattori, M., Sakaki, Y., and Sasaki, H. (2005). Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2–H19 region. Genome Res. 15, 154–165.
| Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2–H19 region.Crossref | GoogleScholarGoogle Scholar | 15590938PubMed |
Yokomine, T., Hata, K., Tsudzuki, M., and Sasaki, H. (2006). Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet. Genome Res. 113, 75–80.
| Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 16575165PubMed |