A quantum leap in avian biology
Leo Joseph A D and Katherine L. Buchanan B CA Australian National Wildlife Collection, CSIRO National Research Collections Australia, GPO Box 1700, Canberra, ACT 2601, Australia.
B Centre for Integrative Ecology, School of Life and Environmental Sciences, Deakin University, Locked Bag 20000, Geelong, Vic. 3217, Australia.
C Editor, Emu – Austral Ornithology.
D Corresponding author. Email: Leo.Joseph@csiro.au
Emu 115(1) 1-5 https://doi.org/10.1071/MUv115n1_ED
Published: 9 February 2015
Friday 12 December 2014 was a red-letter day in the history of ornithology. It saw simultaneous publication of some 27 papers in eight journals, based on a singularly far-reaching and exciting avian dataset: genomic data from 48 species of birds from 32 of the 35 recently proposed avian orders, chosen to span the evolutionary diversity of the class Aves (Zhang et al. 2014a, 2014b, 2014c; Jarvis et al. 2014a, 2014b). The body of work, representing output from more than 200 researchers working in 80 laboratories in 20 countries, reaches almost every corner of ornithology. A flagship paper (Jarvis et al. 2014a) stands on the shoulders of earlier and contemporary work (Hackett et al. 2008; Burleigh et al. 2015) to achieve a nearly complete understanding of relationships among the world’s major avian groups and the timing of the major events in their evolution.
A listing of 20 of the papers associated with the dataset is available at http://www.sciencemag.org/content/346/6215/1308/suppl/DC1 (verified 12 January 2015). One suite of papers, most of which comprise a one-page commentary as well as the full paper, examines the detailed molecular evolution that has made birds the feathered, toothless wonders that they are (Cui et al. 2014; Greenwold et al. 2014; Lovell et al. 2014; Meredith et al. 2014; Romanov et al. 2014; Weber et al. 2014a, 2014b; Wirthlin et al. 2014; Zhang et al. 2014c). Another paper thoroughly examines neontological (from living birds) and palaeontological data to reassess the origins of birds (Xu et al. 2014). Next, a series of papers explores the same data to revolutionise other areas of ornithology: the conservation of endangered species (Callicrate et al. 2014; S. Li et al. 2014), the molecular basis of the responses of birds to long-term global climate change (C. Li et al. 2014), the evolution of sex chromosomes in birds (Zhou et al. 2014), and the neurobiology of song and vocal learning (Pfenning et al. 2014; Whitney et al. 2014). Notably, the entirety of the work published on 12 December 2014 rested on two foundations: museum collections of birds often made with no particular view to future uses of individual specimens (Kress 2014) and the need to invent new analytical methods to handle the volume of data involved (Aberer et al. 2014; B. Li et al. 2014; Mirarab et al. 2014a, 2014b, 2014c). Analyses for Jarvis et al. (2014a), conducted on nine supercomputers, took the equivalent of more than 400 years of single-processor computing.
A genome is the full complement of DNA sequences in an organism. It represents the code for transcription and production of the proteins that build and define the organism’s phenotype. But the genome is not a homogeneous chunk of DNA evolving at one rate under one evolutionary force. It is more an evolutionary patchwork of many different kinds of DNA sequences. Some parts of the genome code for proteins and have intricate rates of evolution. Other portions do not code for proteins and evolve at other rates. Other non-coding DNA sequences are interspersed between genes, as well as within them, the latter being called introns. Some DNA sequences move around the genome, inserting themselves into new positions within it. All contribute to a molecular ‘footprint’ of the evolutionary history of the organism carrying the DNA. The range of evolutionary forces that cause DNA sequences to evolve – that is, to change – includes natural selection and the random process of genetic drift among others. This spectrum of DNA sequences, and their rates of evolution, can be drawn on to answer questions at different evolutionary depths from populations within a species down to commonalities between, say, animals and plants. The more kinds of DNA sequences one can look at, generally, the better are the chances of answering a question using information in DNA.
Given all this, we can now appreciate what has and has not been done in this new body of work. It has not sequenced entire bird genomes from the beginning to end of every chromosome on each of the 48 birds. For now, this is just still too complicated in most complex multicellular organisms. The new work has used new methods from the toolbox of genomics to sample far (and we really mean ‘far’) more of the genomes. After obtaining genomic data from each of the 48 birds, it has assembled and aligned ~41.8 × 106 base pairs (‘rungs’ in the ladder-shaped molecule of DNA (bp)) from 8251 protein-coding genes, also non-coding introns from within 2516 of these genes and a set of 3679 ‘ultraconserved elements’, a recently discovered kind of non-coding DNA of potentially great utility at a range of taxonomic depths (Smith et al. 2014; Crawford et al. 2015). Before 12 December 2014, the most detailed study of relationships among the world’s birds, by Hackett et al. (2008), had looked at ~32000 bp from 19 different pieces of DNA in 169 species.
The papers from 12 December 2014 hint of more to come but a summary of a few main points follows.
Avian phylogeny
Living birds fall into two main groups: the Palaeognathae (ratites, tinamous) and the Neognathae, which comprises the Galloanseres (waterfowl, landfowl) and the Neoaves (all other birds). Problems of determination of the relationships have been most trenchant within the Neoaves because its diversity evolved fairly quickly. This has led to mainly comb-like, rather than tree-like, branching patterns of relationships, the so-called ‘wall of death’ in earlier studies. The analytical power of data from many genes is then needed to discern branching patterns. The new work has delivered this. It has derived an almost complete hypothesis of the branching pattern of relationships among birds and proposes solutions to several contentious issues of relationships within the Neoaves. An example has simply been determining its major component groups, especially among landbirds. The new work argues that the Neoaves comprises the Passerea and the Columbea (Fig. 1). The Columbea has just the mesites of Madagascar, the sandgrouse, pigeons and doves, and flamingos and grebes. All other Neoaves make up the Passerea – including the ever mysterious Hoatzin (Opisthocomus hoazin) of South America as sister (i.e. closest living relative) to plovers and cranes and not closely related to cuckoos elsewhere in the Passerea. Core landbirds within the Passerea, themselves known as Telluraves, comprise two main groups: the Australaves (all passerines, parrots, falcons, and seriemas), and the Afroaves (comprising the redefined Coraciiformes (kingfishers and relatives), woodpeckers, hornbills, trogons, cuckoo-rollers, mousebirds, owls, eagles and New World vultures). These groups had earlier been named after southern hemisphere regions (Ericson 2012), but Jarvis et al. (2014a) stress that wider taxon sampling is needed to address the biogeographical justification of the names Australaves and Afroaves. This should not be confused with the still solid case for Australo-Papuan origins of oscine passerines within the passerines, nor, probably, a southern origin of passerines themselves, all of which are in the Australaves. Nonetheless, a key role for the southern hemisphere in the evolution of the Neoaves is intimated. Jarvis et al. (2014a) conclude that today’s diversity of Neoaves evolved rapidly in evolutionary terms, the divergence between Passerea and Columbea having originated between ~67 and 69 million years ago (Mya) and most ordinal diversity within those groups having evolved by 50 Mya. Much of this evolution is suggested, in turn, as a response to a ‘release of ecological niches following the environmental destruction and species extinctions linked to an asteroid impact’ (Jarvis et al. 2014a), that impact being the one that formed the Chicxulub crater at around 66 Mya (see Benton 2010).
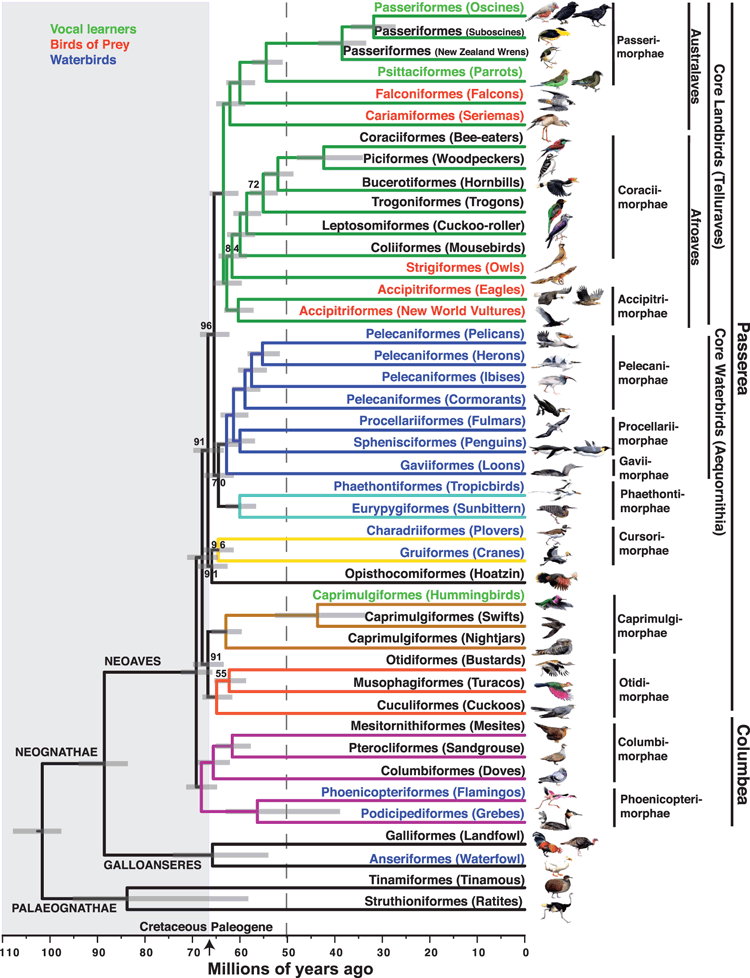
|
Another contentious issue has centred on understanding how convergent evolution has operated, so that true relationships can be discerned from parallel radiations. That is, how does one disentangle convergent similarities owing to a shared life history (such as predation or aquatic life), the suites of morphological and biological traits that have evolved with those life histories, and actual relationships? We now can better see the patterns. Consider birds having life histories centred on aquatic environments. Flamingos and grebes are now very well established as closest relatives (Mayr 2008) within the Columbea. Conversely in the Passerea, a single clade within the core waterbirds (Aequornithia) comprises pelicans, herons, ibises, cormorants, ‘tube-nosed’ seabirds, penguins, loons, and the Sunbittern (Eurypyga helias), another puzzling bird of the Neotropics, as sister to the tropicbirds. Shorebirds and cranes are closest relatives elsewhere within the Passerea. Next, consider raptorial life history and its accompanying morphology. This occurs in falcons on one hand and owls, eagles and New World Vultures on the other. Here the strange seriemas of the Neotropics emerge as the closest relative of falcons, parrots and passerines. This suggests that that particular clade, if not all core landbirds (Telluraves, as above) evolved from an apex predator and involved two losses of raptorial traits. Finally, vocal learning and its neurobiological genetic basis has evolved convergently in hummingbirds, parrots, oscine passerines and, as the new work shows (Pfenning et al. 2014), humans.
Interestingly, when the analyses were conducted on a subset of the data comprising only protein-coding genes (Fig. 4d in Jarvis et al. 2014a), species of similar life-history aligned together, in contrast to the actual pattern of relationships inferred from all of the data. Given the validity of the analysis itself, this raises the questions of whether this problem will be peculiar to birds or whether it suggests caution for other similarly broad-scale phylogenomic studies of other major groups of animals using only protein-coding genes (e.g. Misof et al. 2014).
Molecular level characteristics of birds
Lovell et al. (2014) show that birds lack ~274 protein-coding genes that are present in the genomes of most other vertebrates. These genes are located in regions associated with chromosomal rearrangements, and are largely present in crocodiles (Crocodilia), suggesting that their loss occurred subsequent to the split of dinosaurs and birds from crocodilians. The reasons underlying these genomic omissions are not clear but it has been suggested that this may in part be due to adaptations to flight and enhanced capacity to deal with viral infection. Indeed, many of the missing genes are associated with lethality in rodents, human genetic disorders or biological functions targeting various tissues.
Zhang et al. (2014c) also address the question of why the avian genome is so much smaller than that of mammals (except bats (Chiroptera)). They suggest that it may relate to a capacity for more rapid regulation of genes required during powered flight (as distinct from gliding). Noting also the hollow bones of birds, they observe that of 89 genes involved in ossification, 49 showed evidence of positive selection, and this is substantially higher than the 31 so identified in mammals. Most of these are involved in how processes of bone ossification are regulated. Zhang et al. (2014c) conclude that this kind of regulation may underpin most of the evolutionary remodelling of bones that has led to the structural differences in bone between birds and mammals.
Conservation genomics
S. Li et al. (2014) examined the genomes of the endangered Crested Ibis (Nipponia nippon), the Little Egret (Egretta garzetta), with which the Crested Ibis common co-occurs, and the genomes of 41 other birds under various degrees of threat. They confirm that loss of genetic diversity and increases in deleterious mutations of protein-coding genes contribute to major genetic issues in the endangered Ibis. Inbreeding and loss of functionally important genes in the Crested Ibis may leave the species more susceptible to other environmental factors, such as long-term climate change, over-hunting and overuse of agrochemicals. Conversely, seemingly positive changes in the recovering population of Crested Ibis include genetic diversity increasing between new populations more rapidly than expected, and changes in some gene families that could potentially be related to surviving extinction or recovery. This work establishes a genome-wide DNA identification platform for molecular breeding and conservation practices to speed the sustainable recovery of endangered species.
Analysis of the demographic history of the Crested Ibis revealed two ancient population bottlenecks and one recent one. Specifically, a precipitous population drop to ~10000 individuals was inferred at the end of the last glaciations (0.01 Mya). The more recent event led to a drastic population reduction throughout the last 100 years, until ~25 years ago when human assisted recovery efforts began. This contrasts with the Little Egret, for which population size decreased during the glaciation at 0.3–0.13 Mya but only slightly decreased in the last glaciation.
In effect, S. Li et al. (2014) have thrown a challenge to researchers of endangered birds. Genomic tools are available for studying an endangered study species along with a co-occurring, closely related yet more common species to search for genome-wide signatures of the causes and consequences of rarity and decline.
Evolution of vocal learning
Vocal learning is a trait shared between three avian classes and humans. Pfenning et al. (2014) test whether independent evolution of vocal learning in three different classes of birds (hummingbirds, parrots and oscine passerines) represents convergent evolution through the same genomic solutions to a common problem. Looking at patterns of gene expression in analogous brain regions, they found similar expression patterns in over 50 genes involved in vocal learning in birds and in humans. This suggests that these are key candidate genes for development and motor control of vocal learning and, moreover, that not only has convergent evolution occurred in brain structuring, but also in molecular expression within these regions. A second paper (Whitney et al. 2014) looks at the gene expression in relation to singing behaviour in oscine songbirds. It shows that even before an individual bird first shows singing behaviour, the brain nuclei involved all have different gene expression profiles. Up to 10% of genes within the genome change their levels of expression in relation to song production but this occurs in anatomically specific and temporal waves. Apparently, the act of song production sets off a chain of gene expression events, which itself controls future song production. This study also suggests the further complication that epigenetic mechanisms (the interaction of a non-DNA molecule with the DNA of a gene to control how genes are expressed) appear likely to play a key role in controlling vocal communication in birds.
Conclusion
One could be forgiven for thinking that the world’s birds, along with convergent evolution, have played a very long-running joke on us ornithologists. We have repeatedly been tricked into going down the road of many a wrong hypothesis about who is related to whom among birds. We once thought that the foot-propelled diving birds – grebes and loons – were each other’s closest relatives but we now know that grebes and flamingos are most closely related, or that many of the passerines of the Australian region are closely related to their northern hemisphere namesakes when they are not, or that owlet-nightjars are closely related to nightjars when they are closest relatives to swifts and hummingbirds. Such is the way of science. One of the breakthroughs from these latest scientific revelations is that we have gradually caught up with the joke by way of those examples and many others. The new work sees us at last being very close to understanding the big picture of avian relationships such that we can embark on a surer evolutionary footing into a new era of comparative avian biology, not just phylogenetic relationships. We await the next instalment of similarly sized datasets tackling relationships among all the families of birds.
References
Aberer, A., Stamatakis, A., Mirarab, S., Warnow, T., and Faircloth, B. C. (2014). Species tree and gene tree inferences using maximum likelihood. Supplementary Material SM4 to ‘Whole-genome analyses resolve early branches in the tree of life of modern birds’. (Jarvis et al. 2014a, Science 346, 1320–1331). Available at http://www.sciencemag.org/content/346/6215/1320/suppl/DC1 [Verified 12 January 2015].Benton, M. J. (2010). The origins of modern biodiversity on land. Philosophical Transactions of the Royal Society of London – B. Biological Sciences 365, 3667–3679.
| The origins of modern biodiversity on land.Crossref | GoogleScholarGoogle Scholar |
Burleigh, J. G., Kimball, R. T., and Braun, E. L. (2015). Building the avian tree of life using a large-scale, sparse supermatrix Molecular Phylogenetics and Evolution 84, 53–63.
| Building the avian tree of life using a large-scale, sparse supermatrixCrossref | GoogleScholarGoogle Scholar |
Callicrate, T., Dikow, R., Thomas, J. W, Mullikin, J. C, Jarvis, E. D., Fleischer, R. C., NISC Comparative Sequencing Program (2014). Genomic resources for the endangered Hawaiian honeycreepers. BMC Genomics 15, 1098.
| Genomic resources for the endangered Hawaiian honeycreepers.Crossref | GoogleScholarGoogle Scholar |
Crawford, N. G., Parham, J. F., Sellas, A. B., Faircloth, B. C., Glenn, T. C., Papenfuss, T. J., Henderson, J. B., Hansen, M. H., and Simison, W. B. (2015). A phylogenomic analysis of turtles. Molecular Phylogenetics and Evolution 83, 250–257.
| A phylogenomic analysis of turtles.Crossref | GoogleScholarGoogle Scholar |
Cui, J., Zhao, W., Huang, Z., Jarvis, E. D., Gilbert, M. T. P., Walker, P. J., Holmes, E. C., and Zhang, G. (2014). Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biology 15, 539.
| Low frequency of paleoviral infiltration across the avian phylogeny.Crossref | GoogleScholarGoogle Scholar |
Ericson, P. (2012). Evolution of terrestrial birds in three continents: biogeography and parallel radiations. Journal of Biogeography 39, 813–824.
| Evolution of terrestrial birds in three continents: biogeography and parallel radiations.Crossref | GoogleScholarGoogle Scholar |
Greenwold, M. J., Bao, W., Jarvis, E. D., Hu, H., Cai, L., Gilbert, M. T. P., Zhang, G., and Sawyer, R. H. (2014). Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles. BMC Evolutionary Biology 14, 249.
| Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles.Crossref | GoogleScholarGoogle Scholar |
Hackett, S. J., Kimball, R. T., Reddy, S., Bowie, R. C. K., Braun, E. L., Braun, M. J., Chojnowski, J. L., Cox, W. A., et al (2008). A phylogenomic study of birds reveals their evolutionary history. Science 320, 1763–1768.
| A phylogenomic study of birds reveals their evolutionary history.Crossref | GoogleScholarGoogle Scholar |
Jarvis, E. D., Mirarab, S., Aberer, A. J., Li, B., Houde, P., Ho, S. Y. W., Faircloth, B. C., Nabholz, B., et al (2014a). Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331.
| Whole-genome analyses resolve early branches in the tree of life of modern birds.Crossref | GoogleScholarGoogle Scholar |
Jarvis, E. D., Gilbert, M. T. P., Zhang, G., Howard, J., Fjeldså, J., Jønsson, K. A., Orlando, L., Bertelsen, M., et al (2014b). Rationale for selection of species, sex, tissue sources, and bird collection details. Supplementary Material SM1 to ‘Whole-genome analyses resolve early branches in the tree of life of modern birds’. (Jarvis et al. 2014a, Science 346, 1320–1331). Available at www.sciencemag.org/content/346/6215/1320/suppl/DC1 [Verified 12 January 2015]. , . [Verified 12 January 2015].&title=&date=2014&volume=&spage=&epage=&sid=csiro&aulast=Jarvis&aufirst=E. D." target="_blank" rel="nofollow noopener noreferrer" >

Kress, W. J. (2014). Valuing collections. Science 346, 1310.
| Valuing collections.Crossref | GoogleScholarGoogle Scholar |
Li, B., Jarvis, E. D., Li, C., Zhang, G., Faircloth, B. C., and Mello, C. (2014). Identification and annotation of total evidence nucleotide and whole genome datasets. Supplementary Material SM2 to ‘Whole-genome analyses resolve early branches in the tree of life of modern birds’. (Jarvis et al. 2014a, Science 346, 1320–1331). Available at www.sciencemag.org/content/346/6215/1320/suppl/DC1 [Verified 12 January 2014].
Li, C., Zhang, Y., Li, J., Kong, L., Hu, H.,, Pan, H., Xu, L., Deng, Y., et al (2014). Two Antarctic penguin genomes reveal insights into their evolutionary history and molecular changes related to the Antarctic environment. GigaScience 3, 27.
| Two Antarctic penguin genomes reveal insights into their evolutionary history and molecular changes related to the Antarctic environment.Crossref | GoogleScholarGoogle Scholar |
Li, S., Li, N., Cheng, C., Xiong, Z., Liu, Q., et al (2014). Genomic signatures of near-extinction and rebirth of the Crested Ibis and other endangered bird species. Genome Biology 15, 557.
| Genomic signatures of near-extinction and rebirth of the Crested Ibis and other endangered bird species.Crossref | GoogleScholarGoogle Scholar |
Lovell, P. V., Wirthlin, M., Wilhelm, L., Minx, P., Lazar, N. H., Carbone, L., Warren, W. C., and Mello, C. V. (2014). Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biology 15, 565.
| Conserved syntenic clusters of protein coding genes are missing in birds.Crossref | GoogleScholarGoogle Scholar |
Mayr, G. (2008). Avian higher-level phylogeny: well-supported clades and what we can learn from a phylogenetic analysis of 2954 morphological characters. Journal of Zoological Systematics and Evolutionary Research 46, 63–72.
Meredith, R. W, Zhang, G, Gilbert, M. T. P, Jarvis, E. D., and Springer, M. S. (2014). Evidence for a single loss of mineralized teeth in the common avian ancestor Science 346, 1336.
| Evidence for a single loss of mineralized teeth in the common avian ancestorCrossref | GoogleScholarGoogle Scholar |
Mirarab, S., Warnow, T., Shamsuzzoha Bayzid, Md., Boussau, B., Liu, L., and Edwards, S. (2014a). Species tree inference using multispecies coalescence. Supplementary Material SM5 to ‘Whole-genome analyses resolve early branches in the tree of life of modern birds’. (Jarvis et al. 2014a, Science 346, 1320–1331). Available at www.sciencemag.org/content/346/6215/1320/suppl/DC1 [Verified 12 January 2015].
Mirarab, S., Warnow, T., da Fonseca, R., Gilbert, M. T. P., Li, C., Zhang, G., Faircloth, B. C., and Jarvis, E. D. (2014b). Alignments and their filtering. Supplementary Material SM3 to ‘Whole-genome analyses resolve early branches in the tree of life of modern birds’. (Jarvis et al. 2014a, Science 346, 1320–1331). Available at www.sciencemag.org/content/346/6215/1320/suppl/DC1 [Verified 12 January 2015].
Mirarab, S, Shamsuzzoha Bayzid, Md, Boussau, B, and Warnow, T. (2014c). Statistical binning enables an accurate coalescent-based estimation of the avian tree Science 346, 1250463.
| Statistical binning enables an accurate coalescent-based estimation of the avian treeCrossref | GoogleScholarGoogle Scholar |
Misof, B., Liu, S., Meusemann, K., Peters, R. S., Donath, A., Mayer, C., Frandsen, P. B., Ware, J., et al (2014). Phylogenomics resolves the timing and pattern of insect evolution Science 346, 763–767.
| Phylogenomics resolves the timing and pattern of insect evolutionCrossref | GoogleScholarGoogle Scholar |
Pfenning, A. R., Hara, E., Whitney, O., Rivas, M. V., Wang, R., Roulhac, P. L., Howard, J. T., Wirthlin, M., et al (2014). Convergent transcriptional specializations in the brains of humans and song-learning birds Science 346, 1256846.
| Convergent transcriptional specializations in the brains of humans and song-learning birdsCrossref | GoogleScholarGoogle Scholar |
Romanov, M. N., Farré, M., Lithgow, P. E., Fowler, K. E., Skinner, B. M., O’Connor, R., Fonseka, G., Backström, N., et al (2014). Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genomics 15, 1060.
| Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor.Crossref | GoogleScholarGoogle Scholar |
Smith, B. T., Harvey, M. G., Faircloth, B. C., Glenn, T. C., and Brumfield, R. T. (2014). Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Systematic Biology 63, 83–95.
| Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales.Crossref | GoogleScholarGoogle Scholar |
Weber, C. C., Boussau, B., Romiguier, J., Jarvis, E. D., and Ellegren, H. (2014a). Evidence for GC-biased gene conversion as a driver of between-lineage differences in avian base composition. Genome Biology 15, 549.
| Evidence for GC-biased gene conversion as a driver of between-lineage differences in avian base composition.Crossref | GoogleScholarGoogle Scholar |
Weber, C. C., Nabholz, B., Romiguier, J., and Ellegren, H. (2014b). Kr/Kc but not dN/dS correlates positively with body mass in birds, raising implications for inferring lineage-specific selection. Genome Biology 15, 542.
| Kr/Kc but not dN/dS correlates positively with body mass in birds, raising implications for inferring lineage-specific selection.Crossref | GoogleScholarGoogle Scholar |
Whitney, O., Pfenning, A. R., Howard, J. T., Blatti, C. A., Liu, F., Ward, J. W., Wang, R., Audet, J.-N., et al (2014). Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science 346, 1256780.
| Core and region-enriched networks of behaviorally regulated genes and the singing genome.Crossref | GoogleScholarGoogle Scholar |
Wirthlin, M., Lovell, P. V., Jarvis, E. D., and Mello, C. V. (2014). Comparative genomics reveals molecular features unique to the songbird lineage. BMC Genomics 15, 1082.
| Comparative genomics reveals molecular features unique to the songbird lineage.Crossref | GoogleScholarGoogle Scholar |
Xu, X., Zhou, Z., Dudley, R., Mackem, S., Chuong, C.-M., Erickson, G. M., and Varricchio, D. J. (2014). An integrative approach to understanding bird origins. Science 346, 1253293.
| An integrative approach to understanding bird origins.Crossref | GoogleScholarGoogle Scholar |
Zhang, G., Jarvis, E. D., and Gilbert, M. T. P. (2014a). A flock of genomes. Science 346, 1308–1309.
| A flock of genomes.Crossref | GoogleScholarGoogle Scholar |
Zhang, G., Li, B., Li, C., Gilbert, M. T. P., Jarvis, E. D., Wang, J., The Avian Genome Consortium. (2014b). Comparative genomic data of the Avian Phylogenomics Project. GigaScience 3, 26.
| Comparative genomic data of the Avian Phylogenomics Project.Crossref | GoogleScholarGoogle Scholar |
Zhang, G., Li, C., Li, Q., Li, B., Larkin, D. M., Lee, C., Storz, J. F., Antunes, A., et al (2014c). Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320.
| Comparative genomics reveals insights into avian genome evolution and adaptation.Crossref | GoogleScholarGoogle Scholar |
Zhou, Q., Zhang, J., Bachtrog, D., An, N., Huang, Q., Jarvis, E. D., Gilbert, M. T. P., and Zhang, G. (2014). Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1246338.
| Complex evolutionary trajectories of sex chromosomes across bird taxa.Crossref | GoogleScholarGoogle Scholar |

