Can eDNA be an indicator of tree groundwater use? A perspective
L. Pollitt A C , K. Korbel A , J. Dabovic B , A. Chariton
A C , K. Korbel A , J. Dabovic B , A. Chariton  A and G. C. Hose A
A and G. C. Hose A
A School of Natural Sciences, Macquarie University, Sydney, NSW 2109, Australia.
B Department of Planning and Environment, Water, Parramatta, NSW 2124, Australia.
C Corresponding author. Email: loren.pollitt@hdr.mq.edu.au
Marine and Freshwater Research - https://doi.org/10.1071/MF21293
Submitted: 11 August 2021 Accepted: 25 February 2022 Published online: 12 April 2022
Journal Compilation © CSIRO 2022 Open Access CC BY-NC-ND
Abstract
A major challenge of managing groundwater-dependent ecosystems is determining when and where plants are accessing and using groundwater. Addressing this knowledge gap is particularly pertinent where remnant stands of old growth trees reside within areas where groundwater is being used at an unsustainable rate. The aim of this paper is to investigate what it means to find tree DNA in the groundwater and provide a perspective on whether the detection of tree DNA in groundwater could provide an indicator of groundwater use by trees. This idea arose from recent DNA-based surveys that routinely detected tree DNA in groundwater samples, which may be unexpected given the general absence of plants in dark, subsurface environments. We discuss the likely sources and fate of tree DNA in groundwater and the knowledge needed to progress the development of tree DNA as a robust indicator. If successful, such an indicator would help managers better understand the water requirements of groundwater-dependent vegetation, meet legislative obligations for monitoring and assessment, and improve the conservation and management of groundwater-dependent ecosystems.
Keywords: environmental DNA, riparian vegetation, phreatophytes, groundwater-dependent ecosystems, GDE.
Introduction
The development of next-generation sequencing and its application to environmental monitoring and assessment over the past decade has increased exponentially the range of biota that can be routinely identified. In many studies, researchers collect data on the presence of taxa that are extraneous to the research or monitoring questions. In some cases, these non-target taxa will be those that, although inhabiting the ecosystem, are not of direct interest, or may be taxa that occur outside the ecosystem and may be accidental, invasive or contaminants (Chariton et al. 2015). So, what can we learn from unexpected or ‘non-target’ taxa in environmental DNA (eDNA) samples?
Environmental-DNA metabarcoding (i.e. the identification of multiple species from a single environmental sample (e.g. soil and water) containing complex and degraded DNA) has become a popular tool for examining biodiversity, because it is cost effective, non-invasive, and has high detection probabilities for low-abundance taxa (Taberlet et al. 2012; Bohmann et al. 2014; Ruppert et al. 2019). Traditional field methods for surveying richness and abundance, such as mark–recapture and transect surveys, are often affected by disturbance, errors in taxonomic identification and destruction of habitat (Deiner et al. 2017; Ruppert et al. 2019). eDNA can be used to complement traditional sampling because it has the ability to detect the presence of biota through DNA shed in the environment, which provides a method for sampling difficult-to-access habitats (e.g. subterranean and deep-sea environment). Analysing eDNA also increases the potential of detecting rare and cryptic species as well as increasing taxonomic resolution and accuracy (Jane et al. 2015; Deiner et al. 2017; Taberlet et al. 2018; Ruppert et al. 2019; Nørgaard et al. 2021). In addition to characterising the structures of communities, eDNA has a wide range of other applications, and can be used for examining population dynamics, such as detecting non-indigenous species, and for wildlife DNA forensics, monitoring ecosystem health, and functional profiling (Dejean et al. 2011; Díaz-Ferguson and Moyer 2014; Chariton et al. 2015; Goldberg et al. 2015; Barnes and Turner 2016; Pawlowski et al. 2018; Coble et al. 2019; Beng and Corlett 2020).
Despite the technological advances and increases in the use of eDNA for species detection and conservation, many challenges and limitations remain (Stoeckle et al. 2017). These may arise during field sampling, laboratory processing and the interpretation and analysis of results (Thomsen and Willerslev 2015; Zinger et al. 2016, 2019; Trebitz et al. 2017). Specifically, the limitations can include the use of inappropriate bioinformatic pathways, limited reference databases for taxonomic groups, suboptimal primers and polymerase chain reaction (PCR) conditions, contamination from in the field or during laboratory processing and difficulty in estimating longevity of DNA and its persistence in environments (see sections below; Thomsen and Willerslev 2015; Trebitz et al. 2017; Ruppert et al. 2019; Coble et al. 2019; Saccò et al. 2022).
Recent surveys of shallow alluvial aquifers at 122 sites across arid and semi-arid western New South Wales (NSW), Australia, targeting the V7 region of the 18S rDNA gene have identified diverse eukaryote assemblages in the groundwater (Korbel et al. 2017; Nelson 2020; Pollitt 2020). Biota targeted in these studies included invertebrates and fungi, which are key components of groundwater ecosystems. However, as with many eDNA studies using ‘universal’ primers (i.e. those that amplify very common conserved gene regions that capture a wide range of taxa), samples contained a complex mixture of possibly degraded DNA from various organisms in the ecosystem (Taberlet et al. 2018). These samples included several non-target species, i.e. taxa that are not of interest, which are generally removed from the dataset before processing. Of particular interest was the number of plant species detected in these groundwater samples, which may be unexpected given that functioning photosynthetic organisms are typically absent in the dark subsurface environment (Humphreys 2006). However, for numerous plant species, access to groundwater is critical to meet some or all life-cycle stages (Eamus et al. 2006) and, consequently, it is reasonable that the DNA from such species is detected in groundwater.
Knowing when and where plants are accessing and using groundwater is key to managing and conserving terrestrial groundwater-dependent ecosystems (GDEs), particularly in the face of increasing competition for groundwater resources in a changing climate (Eamus et al. 2006). This is a critical and urgent challenge because groundwater overuse is a major global problem (Wada et al. 2010). In Australia, groundwater accounts for one third of the water used and demand is increasing (Harrington and Cook 2014). In arid and semi-arid regions of Australia, groundwater is often the only reliable source of water and the allocation of water to meet agricultural, environmental, industrial and societal needs is a constant point of contention (Leblanc et al. 2012).
The Murray–Darling Basin (MDB) is the primary region of agricultural production in Australia. It provides over 50% of the nation’s agricultural produce, while using over 70% of Australia’s total water usage (Australian Bureau of Statistics 2019). Much of the MDB has been cleared of native trees to accommodate agriculture (Walker et al. 1993), and trees that remain may be under stress and populations of some species are in decline (Ngugi et al. 2021). Given the immense value of this region, natural resources, such as vegetation and water, need to be carefully monitored. However, the water requirements of the natural environment are often secondary to agricultural and industrial needs (Murray–Darling Basin Authority 2017). The picture in the MDB reflects a global trend, with GDEs being increasingly threatened by groundwater abstraction and declining water tables (Brown et al. 2010; Hoogland et al. 2010; Tomlinson and Boulton 2010; Eamus et al. 2015; Pérez Hoyos et al. 2016; van Engelenburg et al. 2018; Erostate et al. 2020). Tools that provide cost-effective and rapid indication of groundwater dependence are needed globally.
Traditional approaches to determining tree groundwater use include stable isotope analysis of tree and groundwater, sap-flow analysis, and remote sensing, which largely rely on the inference between plant attributes (water flow, greenness) and water use and require specialised equipment or analytical capabilities (Eamus et al. 2006; Cook and Eamus 2018; Cleverly et al. 2020). In comparison to these traditional measurements, eDNA potentially provides a real indicator of plant interactions with groundwater. The aim of this discourse is to consider what does it mean to find tree DNA in the groundwater, and can this be an indicator that trees are accessing groundwater?
Potential sources of plant DNA in groundwater
Plant DNA detected in samples of groundwater could be derived from (1) contamination during collection as groundwater is drawn from a well or spring, (2) DNA leached from surficial plant material through the soil to the groundwater or (3) DNA released from tree roots that are accessing the groundwater and thus submerged in the groundwater (Fig. 1; Groom et al. 2000; Zencich et al. 2002; Poté et al. 2003, 2009a; Bravo et al. 2010; Froend and Sommer 2010). It is important to determine the source of DNA in groundwater if we are to indicate plant–groundwater dependency. Typically, non-woody plants rely on soil moisture unless the groundwater is near to the surface, and it is only woody plants (tree species) with extensive root systems that are able to access groundwater that is several metres or more below the surface (Eamus et al. 2015). It is the detection of such trees that provides potential for the indication of groundwater dependency.
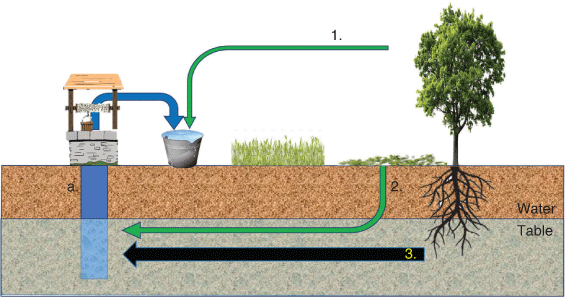
|
A contaminant or the real deal?
There are many challenges that arise from analysing the large datasets created from DNA sequencing. One such challenge is interpreting whether DNA is truly originating from the sample or is potentially reflective of contamination. The presence of ‘unexpected’ DNA in any sample is a possible indicator of contamination and managing and avoiding contamination is important at all stages of the eDNA analysis pipeline (Rees et al. 2015; Ruppert et al. 2019).
In the case of tree DNA in groundwater, contamination is a possible source, because plant material (particularly pollen) may be blown into sampling containers during sample collection or during downstream processing, but this can be managed by meticulous attention to sampling protocols (Gulden et al. 2005; Rosi-Marshall et al. 2007; Thomsen and Willerslev 2015). Furthermore, field-derived contamination should be infrequent, and regular detection of unexpected taxa can provide evidence to support it being a true occurrence. Sampling protocols such as purging bores before collecting samples can remove the likelihood of localised contamination from within the bore hole (Korbel et al. 2017). In the case of vegetation, contamination from sources such as pollen may be seasonal, corresponding with flowering periods or weather conditions at the time of sampling that might increase the likelihood of airborne contamination. Sampling bores on multiple occasions during different seasons may help account for such contamination and would provide further support and greater confidence that the species detection is ‘real’ (de Vet et al. 2009; Ceccherini et al. 2009; Zinger et al. 2016).
During laboratory and bioinformatic processes, contamination and misidentification may result in the identification of ‘unexpected’ DNA; however, this can be mitigated through sterile laboratory conditions and the separation of pre- and post-PCR products (Goldberg et al. 2016; Furlan et al. 2020). Amplicon sequencing errors, poor taxonomic resolution, poor taxonomic match and reference database errors may also provide contamination, errors and the misidentification of DNA (Coissac et al. 2012; Ficetola et al. 2015; Furlan et al. 2020). However, these can be attenuated during library preparation by various bioinformatic approaches, as well as by filtering data with the aid of positive and negative controls (Alsos et al. 2018; Harper et al. 2019; Carøe and Bohmann 2020; Furlan et al. 2020; Gillmore et al. 2021).
Leaching of surface material
The accumulation of plant material on the soil surface, and the subsequent breakdown and leaching of that material through the soil profile to the water table, is also a potential source of plant-derived eDNA in groundwater (Fig. 1). In leaf litter, DNA can degrade rapidly, with persistence in soil likely to be affected by both the species present and water saturation, with terrestrial plant-derived eDNA degrading as quickly as in 2 days (Poté et al. 2005), or persisting for up to 1 year (Gulden et al. 2005; Meier and Wackernagel, 2003). The fate of eDNA transported to groundwater as a leachate is less well known.
Leaching of DNA through the soil profile has been demonstrated for several plant species (e.g. Poté et al. 2007, 2009a, 2009b). The mobilisation of extracellular DNA or its associated leaf material is then dependent on soil hydraulics, including the rate of water flow through the soil, grain size and porosity, and the mineral and organic composition of the soil (Gulden et al. 2005), and also depends on the size (and hence, state of degradation) of the eDNA fragment itself (Turner et al. 2014; Barnes and Turner 2016).
The process of leaching, including adsorption of DNA to soil and organic matter and microbial breakdown, means that DNA fragments reaching groundwater will be degraded. Gulden et al. (2005) suggested that the degradation of DNA in leachate water is most likely to be enzymatic, with a half-life between 2 and 30 h for corn and soybean plants. However, degradation rates are strongly and positively correlated with temperature, water chemistry and fragment length (Jo and Minamoto 2021; Saito and Doi 2021a). The extent of degradation also depends on retention time in the soil, such that groundwater depth is likely to influence the extent of degradation. Groundwater closer to surface or within the topsoil horizon is more likely to contain less degraded DNA than is deeper groundwater, which is reflected in the concentrations of DNA in groundwater often decreasing with depth (Nielsen et al. 2007; Eamus 2009).
Degradation of plant-derived eDNA in the soil profile may potentially be correlated with a reduction in the fragment length. If so, this suggests that fragment length of plant DNA retrieved in groundwater samples could be used to infer the DNA source as leached (if short) or being derived from plants accessing vegetation (if long). Overall, the contributions of DNA from surface leaching may be small when considering the vertical soil-leaching pathway and the potential for rapid microbial and chemical degradation of the DNA in that process (Poté et al. 2003, 2009a; Gulden et al. 2005).
DNA from deep plant roots
The most likely source of tree DNA in groundwater is from tree roots that have grown deeper than the water table. In this case, release of root cells or lysis of root material can release plant DNA (either intra- or extracellular) directly into the groundwater. Deep-rooted tree species in this region are dominated by the Myrtaceae and Mimosaceae, including Eucalyptus camdulensis, E. largiflorens, E. coolabah, Acacia stenophylla. Other common tree species include Callitris endlicheri (Cupressaceae) and Casuarina cunninghamiana (Casuarinaceae). Eucalyptus is by far the most abundant genus and those trees are known for their fast and deep root growth (Benson 2008; Christina et al. 2017), leading to an average rooting depth of ~10 m, and up to 60 m for some species (Stone and Kalisz 1991). If we assume that trees with roots in groundwater release DNA to that environment, then there should be a relationship between the groundwater depth, tree root depth and the detection of tree DNA. Therefore, the detection of tree DNA should identify sites where the tree root zone is within the groundwater table (Eamus et al. 2015).
Fate and persistence of DNA in groundwater
Understanding the longevity, fate and transportation of tree-derived DNA is key to being able to use the presence of that DNA as an indicator of groundwater use. Several factors, including water quality, temperature and UV radiation, are important factors influencing DNA persistence (Zhu 2006; Barnes et al. 2014; Strickler et al. 2015) as is the length of DNA fragment (Deagle et al. 2006). Temperature is considered one of the most critical variables affecting DNA detection and longevity within aquatic environments (Strickler et al. 2015), with DNA being markedly more persistent in cold climates (Zhu 2006; Barnes et al. 2014). Additionally, salinity and pH affect DNA degradation; DNA longevity is greatest in neutral and slightly alkaline environments (Strickler et al. 2015), whereas high salinity increases DNA degradation (Barnes et al. 2014; Collins et al. 2018; Saito and Doi 2021b). Fragment length also appears important for DNA degradation, with shorter fragments usually slower to degrade (Deagle et al. 2006). For example, fragments of 300–400 bp persisted in water for up to 1 week in controlled environments (Dejean et al. 2011), with smaller fragments detected for up to 1 month (Zhu 2006). However, microbial enzymatic degradation of the DNA fragments is a key mechanism. The breakdown of DNA under sterile conditions or environments with low microbial activity is much slower than under natural conditions in surface waters where the half-life may be as low as 2 h (Matsui et al. 2001; Gulden et al. 2005; Zhu 2006; Jo and Minamoto 2021; Saito and Doi 2021a). Fortunately, the low microbial biomass and activity expected in groundwater (see Griebler and Lueders 2009) suggest that DNA may be more persistent in groundwater than in surface waters.
The degradation of DNA has been shown to be slow in groundwater (Poté et al. 2003, 2007, 2009a). This may be due to the stability of groundwater ecosystems in terms of temperature and water quality (Humphreys, 2006), lack of UV-induced DNA degradation, and low microbial biomass and activity (Griebler and Lueders 2009). All of these factors potentially increase DNA persistence when compared with surface environments, although studies in groundwater have been limited (e.g. Zhu 2006) and have detected plasmid DNA in groundwater microcosms with average durations of 48–96 h. Preliminary evidence from cave environments suggest DNA persistence for over 1 month (Boulton et al., in press).
DNA longevity in aquatic environments has mainly been studied using animal species in surface waters. Such studies have indicated that species and life stage affect DNA fragment length, longevity and detectability in contemporary environmental DNA samples, ranging from days to weeks (Dejean et al. 2011; Thomsen et al. 2012a, 2012b; Barnes et al. 2014). However, there have been fewer studies investigating detection and persistence of plant DNA in aquatic environments (Anglès d’Auriac et al. 2019), with Bravo et al. (2010) indicating that the half-life of plant DNA in aquatic sediments was more than 1 day, whereas DNA of plant pathogens in soils is detectable for over 1 year (Kunadiya et al. 2021). The soil and sediment matrix can influence the preservation and absorption of DNA, with the clay and sand content being shown to have a positive effect on the absorption and protection of the DNA from nuclease degradation (Poté et al. 2003; Gulden et al. 2005). However, large knowledge gaps remain in the transport, longevity and degradation rates of DNA in groundwaters (Dale et al. 2002), with no studies having been conducted on plant DNA within these environments.
Knowledge on DNA transportation and retention within groundwaters is important if DNA from plants is to be used as an indicator. In surface waters, DNA can be detected kilometres from its origin (Barnes and Turner 2016; Wacker et al. 2019), although the concentration of the detected DNA decreases with distance (Deiner and Altermatt 2014). This is of benefit in detecting vegetation within groundwater because opportunities to access and collect groundwater are limited to caves, wells and springs, which are often not located immediately adjacent to the sites or vegetation of interest (Larned 2012). Like streams and rivers, groundwaters flow (albeit slowly) such that the water collected at a site reflects its interactions with the environment and biota in areas higher up the hydraulic gradient (i.e. ‘upstream’). Consequently, detection of tree-derived eDNA in groundwater requires that the DNA persist in the environment for a sufficient period for it to be transported to the point of collection.
Groundwater flow rates (i.e. hydraulic conductivity), at least in shallow alluvium of the MDB, range from 10–4 to 102 m day–1 (Bioregional Assessment Programme 2016). Given that DNA may remain detectable in aquatic environments for up to several days, DNA released from trees directly into groundwater may be detected in groundwater abstracted from a bore or well within a radius of up to several hundred metres down the hydraulic gradient (i.e. ‘downstream’) from the tree, but this will depend heavily on the hydrogeology of the aquifer (Jane et al. 2015; Pang et al. 2020).
What do we need to know to make this a useful indicator?
For eDNA to be an effective and reliable method for inferring plant groundwater use, the detection of tree DNA in the groundwater, when present, needs to be consistent, thus avoiding false negative results. From the 122 sites sampled across western NSW (Pollitt 2020) that were sampled more than once, 63 had plant DNA detected in at least one, but not all samples, which suggests a large amount of heterogeneity in the detection of plant DNA. In part, this heterogeneity of detection may be an issue with primer specificity and variation in primer affinity. Because the DNA detected in the groundwater is likely to be degraded, the use of large generic primers that are not designed to specifically target plants (such as the V7 18S rDNA eukaryote primer) may limit the detection of possibly degraded, much shorter DNA fragments and may also influence the taxonomic resolution in the output (Taberlet et al. 1991; Hadziavdic et al. 2014). The use of a short highly conserved gene region for plants may help limit some of the heterogeneity detected. The trnL (UAA) intron, and more specifically the P6 loop of the trnL intron, and the second internal transcribed spacer (ITS2) were identified as good target regions for degraded plant DNA because they are variable, highly conserved and short (Taberlet et al. 2007, 2018; Hollingsworth et al. 2011; Moorhouse-Gann et al. 2018). Because no single barcode is without drawbacks or fits the needs perfectly, future analyses should consider using multiple coding regions with additional markers or barcodes (Hollingsworth et al. 2011). In conjunction with using multiple coding regions, a combination of primers that target short, degraded DNA and longer regions may be beneficial to capture as much of the heterogeneity as possible (Ruppert et al. 2019).
Polymerase chain reaction bias may also contribute to the heterogeneity of the species detected in eDNA samples, in which certain sequences may or may not be amplified. Like other detection technologies, eDNA is not without its imperfections and is susceptible to Type I (false positive) and Type II (false negative) errors (Ficetola et al. 2015; Evans et al. 2017; Lopes et al. 2020). False positives can result from (1) contamination, (2) incorrect detection of nontarget species, or (3) detection of DNA from dead organisms in the ecosystem (Darling and Mahon 2011; Rees et al. 2015; Evans et al. 2017). False negatives occur when taxa are present in the ecosystems but not detected and can result from (1) failure to collect DNA in the sample, (2) insufficient assay sensitivity, (3) PCR dropout or (4) lack of viable or degraded DNA (Darling and Mahon 2011; Rees et al. 2015; Evans et al. 2017). The risk of false negatives may be increased in groundwater samples because of the likely degraded state of the DNA (Ficetola et al. 2015). Increasing sample volumes and replication of each stage of analysis (i.e. sample collection, extractions per sample, and amplifications per extraction) may moderate this risk (Darling and Mahon 2011; Rees et al. 2015; Evans et al. 2017). Ficetola et al. (2015) suggested the following three steps to reduce the risk of bias and improve the robustness and reliability: run occupancy models to estimate detection probability, false negatives and true occupancy, evaluate current methods for controlling false negatives and remove ‘uncertain presences’. The characterisation of DNA collected, including concentration and fragment-size distribution, can begin to shed light on several of these issues.
A better understanding of DNA longevity in soils and groundwater ecosystems will help improve our knowledge and understanding of the origin and fate of eDNA in groundwater and the likelihood of its detection over time (Gulden et al. 2005). Further analysis of whether DNA is present in sediments (where it is typically more stable; Poté et al. 2003; Gulden et al. 2005), or in solution, is needed. The expected short-term persistence of tree-derived eDNA in groundwater means that observations will likely reflect recent conditions, and reduces the likelihood of detecting ‘zombie’ DNA (i.e. DNA from dead, rather than living individuals), but further confirmation is required (Baird and Hajibabaei 2012). The use of RNA for detection of recent biotic activity may also be of benefit. Because RNA is produced only by living organisms and degrades quickly (Chimento et al. 2012; Laroche et al. 2017; von Ammon et al. 2019; Wood et al. 2020), detection of RNA could be used as a reliable marker of plant presence and use of groundwater; however, it is more costly, and difficult to sample and preserve in remote locations.
The limited understanding of local hydrogeology at most sampling locations complicates the interpretation of eDNA detections. Groundwater flow directions are difficult to determine at a local scale and often require interpretation of water levels across a region to determine hydraulic gradients. Often such water-level data are difficult to obtain, and groundwater flows cannot be determined from observations of the site or surface features. Knowledge of groundwater flow paths is critical for determining whether trees are upstream or downstream, and, thus, whether eDNA is likely (or not) to be detected in the groundwater. Even when groundwater flow direction is known, aquifer matrix properties such as porosity and dispersivity determine how eDNA maybe dispersed within the aquifer (e.g. the shape and location of the eDNA ‘plume’ downstream of the tree) and whether this intersects with observation points (bores and wells).
Before eDNA can be used as an indicator of plant groundwater use, two key steps remain. First, the reliability of detecting tree DNA in groundwater samples needs to be improved. Through the testing and use of plant-specific primers and the characterisation of detected DNA by analysing the concentration and fragment sizes may help improve issues with PCR bias and heterogeneity. A greater understanding of the transport, longevity and persistence of DNA in groundwater environments will also help improve the reliability of eDNA as an indicator. The use of methods such as DNA spiking may help reduce this knowledge gap. Once the reliability of detecting tree-derived DNA in groundwater samples is confirmed, a critical step remains. That is, linking tree DNA detection to groundwater use by the tree. This step will require linking eDNA analysis with traditional measurements of vegetation groundwater use (see Eamus et al. 2006), such as measurements of sap flow and transpiration and analysis of water isotopes to validate the approach. Once tested and validated against traditional approaches for assessing tree groundwater use, the use of eDNA would enable rapid and broad-scale assessments of tree water use. Indeed, rapid test kits or mobile PCR systems (e.g. Doi et al. 2021) may enable on-site, real-time analyses.
If eDNA can indicate that trees are accessing groundwater, it will provide a new tool for identifying groundwater-dependent vegetation. Further, by confirming that tree DNA in groundwater is sourced from roots means that it can infer the minimum tree root depth, which cannot be determined simply by other means. Targeted testing of eDNA in groundwater as water tables rise and fall (and move in and out of the root zone), or as plants flower or exhibit signs of stress, will also provide detailed knowledge of the timing of groundwater dependence of the vegetation, and identify periods when groundwater resources and levels must be maintained or have reached critical levels. With such knowledge, groundwater abstraction can be more sustainably managed to meet the needs of water users and the environment. Although limitations and challenges still exist, through increased understanding of the longevity and movement of DNA through the groundwater and site-scale hydrology and groundwater flow, eDNA could provide a new insight into tree groundwater use.
Data availability
Data sharing is not applicable as no new data were generated or analysed during this study
Conflicts of interest
Anthony Chariton is an editor for Marine and Freshwater Research but did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Marine and Freshwater Research encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. Jodie Dabovic is an employee of the funding organisation, but this relationship had no influence on the outcomes of the work reported in this paper. The authors have no further conflicts of interest to declare.
Declaration of funding
This research was made possible by funding from the NSW Department of Planning and Environment.
References
Alsos, I. G., Lammers, Y., Yoccoz, N. G., Jørgensen, T., Sjögren, P., Gielly, L., and Edwards, M. E. (2018). Plant DNA metabarcoding of lake sediments: how does it represent the contemporary vegetation. PLoS One 13, e0195403.| Plant DNA metabarcoding of lake sediments: how does it represent the contemporary vegetation.Crossref | GoogleScholarGoogle Scholar | 29664954PubMed |
Anglès d’Auriac, M. B., Strand, D. A., Mjelde, M., Demars, B. O., and Thaulow, J. (2019). Detection of an invasive aquatic plant in natural water bodies using environmental DNA. PLoS One 14, e0219700.
| Detection of an invasive aquatic plant in natural water bodies using environmental DNA.Crossref | GoogleScholarGoogle Scholar | 31299064PubMed |
Australian Bureau of Statistics (2019). Water use on Australian farms, Australia, 2017–2018. (ABS: Canberra, ACT, Australia.) Available at https://www.abs.gov.au/statistics/industry/agriculture/water-use-australian-farms/2017-18#data-download
Baird, D. J., and Hajibabaei, M. (2012). Biomonitoring 2.0: a new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Molecular Ecology 21, 2039–2044.
| Biomonitoring 2.0: a new paradigm in ecosystem assessment made possible by next-generation DNA sequencing.Crossref | GoogleScholarGoogle Scholar | 22590728PubMed |
Barnes, M. A., and Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics 17, 1–17.
| The ecology of environmental DNA and implications for conservation genetics.Crossref | GoogleScholarGoogle Scholar |
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., and Lodge, D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science & Technology 48, 1819–1827.
| Environmental conditions influence eDNA persistence in aquatic systems.Crossref | GoogleScholarGoogle Scholar |
Beng, K. C., and Corlett, R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodiversity and Conservation 29, 2089–2121.
| Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects.Crossref | GoogleScholarGoogle Scholar |
Benson, J. S. (2008). New South Wales Vegetation Classification and assessment: Part 2 plant communities of the NSW South-western Slopes bioregion and update of NSW Western Plains plant communities, version 2 of the NSWVCA database. Cunninghamia 10, 599–673.
Bioregional Assessment Programme (2016). Namoi hydraulic conductivity measurements. Bioregional Assessment Source Dataset. Available at http://data.bioregionalassessments.gov.au/dataset/5f88517d-8154-411d-907f-4e2c2d12a912 [Verified 12 March 2019].
Bohmann, K., Evans, A., Gilbert, M. T. P., Carvalho, G. R., Creer, S., Knapp, M., Douglas, W. Y., and De Bruyn, M. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology & Evolution 29, 358–367.
| Environmental DNA for wildlife biology and biodiversity monitoring.Crossref | GoogleScholarGoogle Scholar |
Boulton, A. J., Bichuette, M. E., Korbel, K., Stoch, F., Niemiller, M. L., Hose, G. C., and Linke, S. (in press). Recent concepts and approaches for conserving groundwater biodiversity. In ‘Groundwater Ecology and Evolution’. (Eds F. Malard, C. Griebler, and S. Retaux.) (Elsevier.)
Bravo, A. G., Wildi, W., and Poté, J. (2010). Kinetics of plant material mass loss and DNA release in freshwater column. Ecotoxicology and Environmental Safety 73, 1548–1552.
| Kinetics of plant material mass loss and DNA release in freshwater column.Crossref | GoogleScholarGoogle Scholar | 20570352PubMed |
Brown, J., Bach, L., Aldous, A., Wyers, A., and DeGagné, J. (2010). Groundwater-dependent ecosystems in Oregon: an assessment of their distribution and associated threats. Frontiers in Ecology and the Environment 9, 97–102.
| Groundwater-dependent ecosystems in Oregon: an assessment of their distribution and associated threats.Crossref | GoogleScholarGoogle Scholar |
Carøe, C., and Bohmann, K. (2020). Tagsteady: a metabarcoding library preparation protocol to avoid false assignment of sequences to samples. Molecular Ecology Resources 20, 1620–1631.
| Tagsteady: a metabarcoding library preparation protocol to avoid false assignment of sequences to samples.Crossref | GoogleScholarGoogle Scholar | 32663358PubMed |
Ceccherini, M. T., Ascher, J., Agnelli, A., Borgogni, F., Pantani, O. L., and Pietramellara, G. (2009). Experimental discrimination and molecular characterization of the extracellular soil DNA fraction. Antonie van Leeuwenhoek 96, 653–657.
| Experimental discrimination and molecular characterization of the extracellular soil DNA fraction.Crossref | GoogleScholarGoogle Scholar | 19533410PubMed |
Chariton, A. A., Stephenson, S., Morgan, M. J., Steven, A. D., Colloff, M. J., Court, L. N., and Hardy, C. M. (2015). Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries. Environmental Pollution 203, 165–174.
| Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries.Crossref | GoogleScholarGoogle Scholar | 25909325PubMed |
Chimento, A., Cacciola, S. O., and Garbelotto, M. (2012). Detection of mRNA by reverse‐transcription PCR as an indicator of viability in Phytophthora ramorum. Forest Pathology 42, 14–21.
| Detection of mRNA by reverse‐transcription PCR as an indicator of viability in Phytophthora ramorum.Crossref | GoogleScholarGoogle Scholar |
Christina, M., Nouvellon, Y., Laclau, J. P., Stape, J. L., Bouillet, J. P., Lambais, G. R., and Le Maire, G. (2017). Importance of deep water uptake in tropical eucalypt forest. Functional Ecology 31, 509–519.
| Importance of deep water uptake in tropical eucalypt forest.Crossref | GoogleScholarGoogle Scholar |
Cleverly, J., Vote, C., Isaac, P., Ewenz, C., Harahap, M., Beringer, J., Campbell, D. I., Daly, E., Eamus, D., He, L., and Hunt, J. (2020). Carbon, water and energy fluxes in agricultural systems of Australia and New Zealand. Agricultural and Forest Meteorology 287, 107934.
| Carbon, water and energy fluxes in agricultural systems of Australia and New Zealand.Crossref | GoogleScholarGoogle Scholar |
Coble, A. A., Flinders, C. A., Homyack, J. A., Penaluna, B. E., Cronn, R. C., and Weitemier, K. (2019). eDNA as a tool for identifying freshwater species in sustainable forestry: a critical review and potential future applications. The Science of the Total Environment 649, 1157–1170.
| eDNA as a tool for identifying freshwater species in sustainable forestry: a critical review and potential future applications.Crossref | GoogleScholarGoogle Scholar | 30308887PubMed |
Coissac, E., Riaz, T., and Puillandre, N. (2012). Bioinformatic challenges for DNA metabarcoding of plants and animals. Molecular Ecology 21, 1834–1847.
| Bioinformatic challenges for DNA metabarcoding of plants and animals.Crossref | GoogleScholarGoogle Scholar | 22486822PubMed |
Collins, R. A., Wangensteen, O. S., O’Gorman, E. J., Mariani, S., Sims, D. W., and Genner, M. J. (2018). Persistence of environmental DNA in marine systems. Communications Biology 1, 185.
| Persistence of environmental DNA in marine systems.Crossref | GoogleScholarGoogle Scholar | 30417122PubMed |
Cook, P. G., and Eamus, D. (2018). The potential for groundwater use by vegetation in the Australian arid zone. Northern Territory Department of Environmental and Natural Resources Water Resources Division. (Northern Territory Government, Darwin, NT, Australia.) https://depws.nt.gov.au/__data/assets/pdf_file/0004/498883/The-Potential-Use-for-Groundwater-Use-by-Vegetation-in-the-Aust.-Arid-Zone.pdf [Verified 15 June 2019].
Dale, P. J., Clarke, B., and Fontes, E. M. (2002). Potential for the environmental impact of transgenic crops. Nature Biotechnology 20, 567–574.
| Potential for the environmental impact of transgenic crops.Crossref | GoogleScholarGoogle Scholar | 12042859PubMed |
Darling, J. A., and Mahon, A. R. (2011). From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environmental Research 111, 978–988.
| From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments.Crossref | GoogleScholarGoogle Scholar | 21353670PubMed |
de Vet, W. W. J. M., Dinkla, I. J. T., Muyzer, G., Rietveld, L. C., and Van Loosdrecht, M. C. M. (2009). Molecular characterization of microbial populations in groundwater sources and sand filters for drinking water production. Water Research 43, 182–194.
| Molecular characterization of microbial populations in groundwater sources and sand filters for drinking water production.Crossref | GoogleScholarGoogle Scholar |
Deagle, B. E., Eveson, J. P., and Jarman, S. N. (2006). Quantification of damage in DNA recovered from highly degraded samples: a case study on DNA in faeces. Frontiers in Zoology 3, 11.
| Quantification of damage in DNA recovered from highly degraded samples: a case study on DNA in faeces.Crossref | GoogleScholarGoogle Scholar | 16911807PubMed |
Deiner, K., and Altermatt, F. (2014). Transport distance of invertebrate environmental DNA in a natural river. PLoS One 9, e88786.
| Transport distance of invertebrate environmental DNA in a natural river.Crossref | GoogleScholarGoogle Scholar | 24523940PubMed |
Deiner, K., Bik, H. M., Mächler, E., Seymour, M., Lacoursière‐Roussel, A., Altermatt, F., Creer, S., Bista, I., Lodge, D. M., De Vere, N., and Pfrender, M. E. (2017). Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Molecular Ecology 26, 5872–5895.
| Environmental DNA metabarcoding: transforming how we survey animal and plant communities.Crossref | GoogleScholarGoogle Scholar | 28921802PubMed |
Dejean, T., Valentini, A., Duparc, A., Pellier-Cuit, S., Pompanon, F., Taberlet, P., and Miaud, C. (2011). Persistence of environmental DNA in freshwater ecosystems. PLoS One 6, e23398.
| Persistence of environmental DNA in freshwater ecosystems.Crossref | GoogleScholarGoogle Scholar | 21858099PubMed |
Díaz-Ferguson, E. E., and Moyer, G. R. (2014). History, applications, methodological issues and perspectives for the use environmental DNA (eDNA) in marine and freshwater environments. Revista de Biología Tropical 62, 1273–1284.
| History, applications, methodological issues and perspectives for the use environmental DNA (eDNA) in marine and freshwater environments.Crossref | GoogleScholarGoogle Scholar | 25720166PubMed |
Doi, H., Watanabe, T., Nishizawa, N., Saito, T., Nagata, H., Kameda, Y., Maki, N., Ikeda, K., and Fukuzawa, T. (2021). On-site environmental DNA detection of species using ultrarapid mobile PCR. Molecular Ecology Resources 21, 2364–2368.
| On-site environmental DNA detection of species using ultrarapid mobile PCR.Crossref | GoogleScholarGoogle Scholar | 34139102PubMed |
Eamus, D. (2009). ‘Identifying groundwater dependent ecosystems: a guide for land and water managers.’ (Land and Water Australia: Sydney, NSW, Australia.)
Eamus, D., Froend, R., Loomes, R., Hose, G., and Murray, B. (2006). A functional methodology for determining the groundwater regime needed to maintain the health of groundwater-dependent vegetation. Australian Journal of Botany 54, 97–114.
| A functional methodology for determining the groundwater regime needed to maintain the health of groundwater-dependent vegetation.Crossref | GoogleScholarGoogle Scholar |
Eamus, D., Zolfaghar, S., Villalobos-Vega, R., Cleverly, J., and Huete, A. (2015). Groundwater-dependent ecosystems: recent insights from satellite and field-based studies. Hydrology and Earth System Sciences 19, 4229–4256.
| Groundwater-dependent ecosystems: recent insights from satellite and field-based studies.Crossref | GoogleScholarGoogle Scholar |
Erostate, M., Huneau, F., Garel, E., Ghiotti, S., Vystavna, Y., Garrido, M., and Pasqualini, V. (2020). Groundwater dependent ecosystems in coastal Mediterranean regions: characterization, challenges and management for their protection. Water Research 172, 115461.
| Groundwater dependent ecosystems in coastal Mediterranean regions: characterization, challenges and management for their protection.Crossref | GoogleScholarGoogle Scholar | 31951946PubMed |
Evans, N. T., Shirey, P. D., Wieringa, J. G., Mahon, A. R., and Lamberti, G. A. (2017). Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 42, 90–99.
| Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing.Crossref | GoogleScholarGoogle Scholar |
Ficetola, G. F., Pansu, J., Bonin, A., Coissac, E., Giguet‐Covex, C., De Barba, M., Gielly, L., Lopes, C. M., Boyer, F., Pompanon, F., and Rayé, G. (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Molecular Ecology Resources 15, 543–556.
| Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data.Crossref | GoogleScholarGoogle Scholar | 25327646PubMed |
Froend, R., and Sommer, B. (2010). Phreatophytic vegetation response to climatic and abstraction-induced groundwater drawdown: examples of long-term spatial and temporal variability in community response. Ecological Engineering 36, 1191–1200.
| Phreatophytic vegetation response to climatic and abstraction-induced groundwater drawdown: examples of long-term spatial and temporal variability in community response.Crossref | GoogleScholarGoogle Scholar |
Furlan, E. M., Davis, J., and Duncan, R. P. (2020). Identifying error and accurately interpreting environmental DNA metabarcoding results: a case study to detect vertebrates at arid zone waterholes. Molecular Ecology Resources 20, 1259–1276.
| Identifying error and accurately interpreting environmental DNA metabarcoding results: a case study to detect vertebrates at arid zone waterholes.Crossref | GoogleScholarGoogle Scholar | 32310337PubMed |
Gillmore, M. L., Golding, L. A., Chariton, A. A., Stauber, J. L., Stephenson, S., Gissi, F., Greenfield, P., Juillot, F., and Jolley, D. F. (2021). Metabarcoding reveals changes in benthic eukaryote and prokaryote community composition along a tropical marine sediment nickel gradient. Environmental Toxicology and Chemistry 40, 1892–1905.
| Metabarcoding reveals changes in benthic eukaryote and prokaryote community composition along a tropical marine sediment nickel gradient.Crossref | GoogleScholarGoogle Scholar |
Goldberg, C. S., Strickler, K. M., and Pilliod, D. S. (2015). Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biological Conservation 183, 1–3.
| Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms.Crossref | GoogleScholarGoogle Scholar |
Goldberg, C. S., Turner, C. R., Deiner, K., Klymus, K. E., Thomsen, P. F., Murphy, M. A., Spear, S. F., McKee, A., Oyler‐McCance, S. J., Cornman, R. S., and Laramie, M. B. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution 7, 1299–1307.
| Critical considerations for the application of environmental DNA methods to detect aquatic species.Crossref | GoogleScholarGoogle Scholar |
Griebler, C., and Lueders, T. (2009). Microbial biodiversity in groundwater ecosystems. Freshwater Biology 54, 649–677.
| Microbial biodiversity in groundwater ecosystems.Crossref | GoogleScholarGoogle Scholar |
Groom, P. K., Froend, R. H., Mattiske, E. M., and Koch, B. L. (2000). Myrtaceous shrub species respond to long-term decreasing groundwater levels on the Gnangara Groundwater Mound, northern Swan Coastal Plain. Journal of the Royal Society of Western Australia 83, 75–82.
Gulden, R. H., Lerat, S., Hart, M. M., Powell, J. R., Trevors, J. T., Pauls, K. P., Klironomos, J. N., and Swanton, C. J. (2005). Quantitation of transgenic plant DNA in leachate water: real-time polymerase chain reaction analysis. Journal of Agricultural and Food Chemistry 53, 5858–5865.
| Quantitation of transgenic plant DNA in leachate water: real-time polymerase chain reaction analysis.Crossref | GoogleScholarGoogle Scholar | 16028966PubMed |
Hadziavdic, K., Lekang, K., Lanzen, A., Jonassen, I., Thompson, E. M., and Troedsson, C. (2014). Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One 9, e87624.
| Characterization of the 18S rRNA gene for designing universal eukaryote specific primers.Crossref | GoogleScholarGoogle Scholar | 24516555PubMed |
Harper, L. R., Buxton, A. S., Rees, H. C., Bruce, K., Brys, R., Halfmaerten, D., Read, D. S., Watson, H. V., Sayer, C. D., Jones, E. P., and Priestley, V. (2019). Prospects and challenges of environmental DNA (eDNA) monitoring in freshwater ponds. Hydrobiologia 826, 25–41.
| Prospects and challenges of environmental DNA (eDNA) monitoring in freshwater ponds.Crossref | GoogleScholarGoogle Scholar |
Harrington, N., and Cook, P. (2014). ‘Groundwater in Australia.’ (National Centre for Groundwater Research and Training: Adelaide, SA, Australia.)
Hollingsworth, P. M., Graham, S. W., and Little, D. P. (2011). Choosing and using a plant DNA barcode. PLoS One 6, e19254.
| Choosing and using a plant DNA barcode.Crossref | GoogleScholarGoogle Scholar | 21637336PubMed |
Hoogland, T., Heuvelink, G. B., and Knotters, M. (2010). Mapping water-table depths over time to assess desiccation of groundwater-dependent ecosystems in the Netherlands. Wetlands 30, 137–147.
| Mapping water-table depths over time to assess desiccation of groundwater-dependent ecosystems in the Netherlands.Crossref | GoogleScholarGoogle Scholar |
Humphreys, W. F. (2006). Aquifers: the ultimate groundwater-dependent ecosystems. Australian Journal of Botany 54, 115–132.
| Aquifers: the ultimate groundwater-dependent ecosystems.Crossref | GoogleScholarGoogle Scholar |
Jane, S. F., Wilcox, T. M., McKelvey, K. S., Young, M. K., Schwartz, M. K., Lowe, W. H., Letcher, B. H., and Whiteley, A. R. (2015). Distance, flow and PCR inhibition: e DNA dynamics in two headwater streams. Molecular Ecology Resources 15, 216–227.
| Distance, flow and PCR inhibition: e DNA dynamics in two headwater streams.Crossref | GoogleScholarGoogle Scholar | 24890199PubMed |
Jo, T., and Minamoto, T. (2021). Complex interactions between environmental DNA (eDNA) state and water chemistries on eDNA persistence suggested by meta‐analyses. Molecular Ecology Resources 21, 1490–1503.
| Complex interactions between environmental DNA (eDNA) state and water chemistries on eDNA persistence suggested by meta‐analyses.Crossref | GoogleScholarGoogle Scholar | 33580561PubMed |
Korbel, K., Chariton, A., Stephenson, S., Greenfield, P., and Hose, G. C. (2017). Wells provide a distorted view of life in the aquifer: implications for sampling, monitoring and assessment of groundwater ecosystems. Scientific Reports 7, 40702.
| Wells provide a distorted view of life in the aquifer: implications for sampling, monitoring and assessment of groundwater ecosystems.Crossref | GoogleScholarGoogle Scholar | 28102290PubMed |
Kunadiya, M. B., Burgess, T. I. A., Dunstan, W., White, D., and Hardy, G. E. S. (2021). Persistence and degradation of Phytophthora cinnamomi DNA and RNA in different soil types. Environmental DNA 3, 92–104.
| Persistence and degradation of Phytophthora cinnamomi DNA and RNA in different soil types.Crossref | GoogleScholarGoogle Scholar |
Larned, S. T. (2012). Phreatic groundwater ecosystems: research frontiers for freshwater ecology. Freshwater Biology 57, 885–906.
| Phreatic groundwater ecosystems: research frontiers for freshwater ecology.Crossref | GoogleScholarGoogle Scholar |
Laroche, O., Wood, S. A., Tremblay, L. A., Lear, G., Ellis, J. I., and Pochon, X. (2017). Metabarcoding monitoring analysis: the pros and cons of using co-extracted environmental DNA and RNA data to assess offshore oil production impacts on benthic communities. PeerJ 5, e3347.
| Metabarcoding monitoring analysis: the pros and cons of using co-extracted environmental DNA and RNA data to assess offshore oil production impacts on benthic communities.Crossref | GoogleScholarGoogle Scholar | 28533985PubMed |
Leblanc, M., Tweed, S., Van Dijk, A., and Timbal, B. (2012). A review of historic and future hydrological changes in the Murray–Darling Basin. Global and Planetary Change 80–81, 226–246.
| A review of historic and future hydrological changes in the Murray–Darling Basin.Crossref | GoogleScholarGoogle Scholar |
Lopes, C. M., Baêta, D., Valentini, A., Lyra, M. L., Sabbag, A. F., Gasparini, J. L., Dejean, T., Haddad, C. F. B., and Zamudio, K. R. (2020). Lost and found: frogs in a biodiversity hotspot rediscovered with environmental DNA. Molecular Ecology 30, 3289–3298.
| 32786119PubMed |
Matsui, K., Honjo, M., and Kawabata, Z. (2001). Estimation of the fate of dissolved DNA in thermally stratified lake water from the stability of exogenous plasmid DNA. Aquatic Microbial Ecology 26, 95–102.
| Estimation of the fate of dissolved DNA in thermally stratified lake water from the stability of exogenous plasmid DNA.Crossref | GoogleScholarGoogle Scholar |
Meier, P., and Wackernagel, W. (2003). Monitoring the spread of recombinant DNA from field plots with transgenic sugar beet plants by PCR and natural transformation of Pseudomonas stutzeri. Transgenic Research 12, 293–304.
| Monitoring the spread of recombinant DNA from field plots with transgenic sugar beet plants by PCR and natural transformation of Pseudomonas stutzeri.Crossref | GoogleScholarGoogle Scholar | 12779118PubMed |
Moorhouse-Gann, R. J., Dunn, J. C., De Vere, N., Goder, M., Cole, N., Hipperson, H., and Symondson, W. O. (2018). New universal ITS2 primers for high-resolution herbivory analyses using DNA metabarcoding in both tropical and temperate zones. Scientific Reports 8, 8542.
| New universal ITS2 primers for high-resolution herbivory analyses using DNA metabarcoding in both tropical and temperate zones.Crossref | GoogleScholarGoogle Scholar | 29867115PubMed |
Murray–Darling Basin Authority (2017). Native vegetation 2017 Basin Plan evaluation. (MDBA: Canberra, ACT, Australia.) Available at https://www.mdba.gov.au/sites/default/files/pubs/BPE-tech-reports-vegetation-2.pdf
Nelson, T. (2020). Factors influencing groundwater microbial communities across an intensive agricultural landscape. M.Res. Thesis, Macquarie University, Sydney, NSW, Australia.
Ngugi, M. R., Neldner, V. J., Dowling, R. M., and Li, J. (2021). Recruitment and demographic structure of floodplain tree species in the Queensland Murray–Darling basin, Australia. Ecological Management & Restoration 23, 64–73.
Nielsen, K. M., Johnsen, P. J., Bensasson, D., and Daffonchio, D. (2007). Release and persistence of extracellular DNA in the environment. Environmental Biosafety Research 6, 37–53.
| Release and persistence of extracellular DNA in the environment.Crossref | GoogleScholarGoogle Scholar | 17961479PubMed |
Nørgaard, L., Olesen, C. R., Trøjelsgaard, K., Pertoldi, C., Nielsen, J. L., Taberlet, P., Ruiz-González, A., De Barba, M., and Iacolina, L. (2021). eDNA metabarcoding for biodiversity assessment, generalist predators as sampling assistants. Scientific Reports 11, 1–12.
Pang, L., Abeysekera, G., Hanning, K., Premaratne, A., Robson, B., Abraham, P., Sutton, R., Hanson, C., Hadfield, J., Heiligenthal, L., and Stone, D. (2020). Water tracking in surface water, groundwater and soils using free and alginate-chitosan encapsulated synthetic DNA tracers. Water Research 184, 116192.
| Water tracking in surface water, groundwater and soils using free and alginate-chitosan encapsulated synthetic DNA tracers.Crossref | GoogleScholarGoogle Scholar | 32731038PubMed |
Pawlowski, J., Kelly-Quinn, M., Altermatt, F., Apothéloz-Perret-Gentil, L., Beja, P., Boggero, A., Borja, A., Bouchez, A., Cordier, T., Domaizon, I., and Feio, M. J. (2018). The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. The Science of the Total Environment 637–638, 1295–1310.
| The future of biotic indices in the ecogenomic era: integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems.Crossref | GoogleScholarGoogle Scholar | 29801222PubMed |
Pérez Hoyos, I. C., Krakauer, N. Y., Khanbilvardi, R., and Armstrong, R. A. (2016). A review of advances in the identification and characterization of groundwater dependent ecosystems using geospatial technologies. Geosciences 6, 17.
| A review of advances in the identification and characterization of groundwater dependent ecosystems using geospatial technologies.Crossref | GoogleScholarGoogle Scholar |
Pollitt, L. (2020). The detection of phreatophytic tree DNA in groundwater, and its environmental correlates. M.Res Thesis, Macquarie University, Sydney, NSW, Australia.
Poté, J., Ceccherini, M. T., Rosselli, W., Wildi, W., Simonet, P., and Vogel, T. M. (2003). Fate and transport of antibiotic resistance genes in saturated soil columns. European Journal of Soil Biology 39, 65–71.
| Fate and transport of antibiotic resistance genes in saturated soil columns.Crossref | GoogleScholarGoogle Scholar |
Poté, J., Rossé, P., Rosselli, W., and Wildi, W. (2005). Kinetics of mass and DNA decomposition in tomato leaves. Chemosphere 61, 677–684.
| Kinetics of mass and DNA decomposition in tomato leaves.Crossref | GoogleScholarGoogle Scholar | 15878608PubMed |
Poté, J., Rosselli, W., Wigger, A., and Wildi, W. (2007). Release and leaching of plant DNA in unsaturated soil column. Ecotoxicology and Environmental Safety 68, 293–298.
| Release and leaching of plant DNA in unsaturated soil column.Crossref | GoogleScholarGoogle Scholar | 17187857PubMed |
Poté, J., Ackermann, R., and Wildi, W. (2009a). Plant leaf mass loss and DNA release in freshwater sediments. Ecotoxicology and Environmental Safety 72, 1378–1383.
| Plant leaf mass loss and DNA release in freshwater sediments.Crossref | GoogleScholarGoogle Scholar | 19419763PubMed |
Poté, J., Mavingui, P., Navarro, E., Rosselli, W., Wildi, W., Simonet, P., and Vogel, T. M. (2009b). Extracellular plant DNA in Geneva groundwater and traditional artesian drinking water fountains. Chemosphere 75, 498–504.
| Extracellular plant DNA in Geneva groundwater and traditional artesian drinking water fountains.Crossref | GoogleScholarGoogle Scholar | 19171370PubMed |
Rees, H. C., Gough, K. C., Middleditch, D. J., Patmore, J. R., and Maddison, B. C. (2015). Applications and limitations of measuring environmental DNA as indicators of the presence of aquatic animals. Journal of Applied Ecology 52, 827–831.
| Applications and limitations of measuring environmental DNA as indicators of the presence of aquatic animals.Crossref | GoogleScholarGoogle Scholar |
Rosi-Marshall, E. J., Tank, J. L., Royer, T. V., Whiles, M. R., Evans-White, M., Chambers, C., Griffiths, N. A., Pokelsek, J., and Stephen, M. L. (2007). Toxins in transgenic crop byproducts may affect headwater stream ecosystems. Proceedings of the National Academy of Sciences of the United States of America 104, 16204–16208.
| Toxins in transgenic crop byproducts may affect headwater stream ecosystems.Crossref | GoogleScholarGoogle Scholar | 17923672PubMed |
Ruppert, K. M., Kline, R. J., and Rahman, M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Global Ecology and Conservation 17, e00547.
| Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA.Crossref | GoogleScholarGoogle Scholar |
Saccò, M., Guzik, M. T., van der Heyde, M., Nevill, P., Cooper, S. J., Austin, A. D., Coates, P. J., Allentoft, M. E., and White, N. E. (2022). eDNA in subterranean ecosystems: applications, technical aspects, and future prospects. The Science of the Total Environment 820, 153223.
| eDNA in subterranean ecosystems: applications, technical aspects, and future prospects.Crossref | GoogleScholarGoogle Scholar | 35063529PubMed |
Saito, T., and Doi, H. (2021a). A model and simulation of the influence of temperature and amplicon length on environmental DNA degradation rates: a meta-analysis approach. Frontiers in Ecology and Evolution 9, 623831.
| A model and simulation of the influence of temperature and amplicon length on environmental DNA degradation rates: a meta-analysis approach.Crossref | GoogleScholarGoogle Scholar |
Saito, T., and Doi, H. (2021b). Degradation modeling of water environmental DNA: experiments on multiple DNA sources in pond and seawater. Environmental DNA 3, 850–860.
| Degradation modeling of water environmental DNA: experiments on multiple DNA sources in pond and seawater.Crossref | GoogleScholarGoogle Scholar |
Stoeckle, B. C., Beggel, S., Cerwenka, A. F., Motivans, E., Kuehn, R., and Geist, J. (2017). A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems. PLoS One 12, e0189119.
| A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems.Crossref | GoogleScholarGoogle Scholar | 29220394PubMed |
Stone, E. L., and Kalisz, P. J. (1991). On the maximum extent of tree roots. Forest Ecology and Management 46, 59–102.
| On the maximum extent of tree roots.Crossref | GoogleScholarGoogle Scholar |
Strickler, K. M., Fremier, A. K., and Goldberg, C. S. (2015). Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation 183, 85–92.
| Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms.Crossref | GoogleScholarGoogle Scholar |
Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17, 1105–1109.
| Universal primers for amplification of three non-coding regions of chloroplast DNA.Crossref | GoogleScholarGoogle Scholar | 1932684PubMed |
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., Vermat, T., Corthier, G., Brochmann, C., and Willerslev, E. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Research 35, e14.
| Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding.Crossref | GoogleScholarGoogle Scholar | 17169982PubMed |
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C., and Willerslev, E. (2012). Towards next‐generation biodiversity assessment using DNA metabarcoding. Molecular Ecology 21, 2045–2050.
| Towards next‐generation biodiversity assessment using DNA metabarcoding.Crossref | GoogleScholarGoogle Scholar | 22486824PubMed |
Taberlet, P., Bonin, A., Coissac, E., and Zinger, L. (2018). DNA metabarcode choice and design. In ‘Environmental DNA: for Biodiversity Research and Monitoring’. pp. 1–5. (Oxford University Press.)
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA: an emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183, 4–18.
| Environmental DNA: an emerging tool in conservation for monitoring past and present biodiversity.Crossref | GoogleScholarGoogle Scholar |
Thomsen, P. F., Kielgast, J., Iversen, L. L., Møller, P. R., Rasmussen, M., and Willerslev, E. (2012a). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS One 7, e41732.
| Detection of a diverse marine fish fauna using environmental DNA from seawater samples.Crossref | GoogleScholarGoogle Scholar | 22952584PubMed |
Thomsen, P. F., Kielgast, J. O. S., Iversen, L. L., Wiuf, C., Rasmussen, M., Gilbert, M. T. P., Orlando, L., and Willerslev, E. (2012b). Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology 21, 2565–2573.
| Monitoring endangered freshwater biodiversity using environmental DNA.Crossref | GoogleScholarGoogle Scholar | 22151771PubMed |
Tomlinson, M., and Boulton, A. J. (2010). Ecology and management of subsurface groundwater dependent ecosystems in Australia: a review. Marine and Freshwater Research 61, 936–949.
| Ecology and management of subsurface groundwater dependent ecosystems in Australia: a review.Crossref | GoogleScholarGoogle Scholar |
Trebitz, A. S., Hoffman, J. C., Darling, J. A., Pilgrim, E. M., Kelly, J. R., Brown, E. A., Chadderton, W. L., Egan, S. P., Grey, E. K., Hashsham, S. A., and Klymus, K. E. (2017). Early detection monitoring for aquatic non-indigenous species: optimizing surveillance, incorporating advanced technologies, and identifying research needs. Journal of Environmental Management 202, 299–310.
| Early detection monitoring for aquatic non-indigenous species: optimizing surveillance, incorporating advanced technologies, and identifying research needs.Crossref | GoogleScholarGoogle Scholar | 28738203PubMed |
Turner, C. R., Barnes, M. A., Xu, C. C., Jones, S. E., Jerde, C. L., and Lodge, D. M. (2014). Particle size distribution and optimal capture of aqueous microbial eDNA. Methods in Ecology and Evolution 5, 676–684.
| Particle size distribution and optimal capture of aqueous microbial eDNA.Crossref | GoogleScholarGoogle Scholar |
van Engelenburg, J., Hueting, R., Rijpkema, S., Teuling, A. J., Uijlenhoet, R., and Ludwig, F. (2018). Impact of changes in groundwater extractions and climate change on groundwater-dependent ecosystems in a complex hydrogeological setting. Water Resources Management 32, 259–272.
| Impact of changes in groundwater extractions and climate change on groundwater-dependent ecosystems in a complex hydrogeological setting.Crossref | GoogleScholarGoogle Scholar |
von Ammon, U., Wood, S. A., Laroche, O., Zaiko, A., Lavery, S. D., Inglis, G. J., and Pochon, X. (2019). Linking environmental DNA and RNA for improved detection of the marine invasive fanworm Sabella spallanzanii. Frontiers in Marine Science 6, 621.
| Linking environmental DNA and RNA for improved detection of the marine invasive fanworm Sabella spallanzanii.Crossref | GoogleScholarGoogle Scholar |
Wacker, S., Fossøy, F., Larsen, B. M., Brandsegg, H., Sivertsgård, R., and Karlsson, S. (2019). Downstream transport and seasonal variation in freshwater pearl mussel (Margaritifera margaritifera) eDNA concentration. Environmental DNA 1, 64–73.
| Downstream transport and seasonal variation in freshwater pearl mussel (Margaritifera margaritifera) eDNA concentration.Crossref | GoogleScholarGoogle Scholar |
Wada, Y., Van Beek, L. P., Van Kempen, C. M., Reckman, J. W., Vasak, S., and Bierkens, M. F. (2010). Global depletion of groundwater resources. Geophysical Research Letters 37, L20402.
| Global depletion of groundwater resources.Crossref | GoogleScholarGoogle Scholar |
Walker, J., Bullen, F., and Williams, B. G. (1993). Ecohydrological changes in the Murray–Darling Basin. I. The number of trees cleared over two centuries. Journal of Applied Ecology 30, 265–273.
| Ecohydrological changes in the Murray–Darling Basin. I. The number of trees cleared over two centuries.Crossref | GoogleScholarGoogle Scholar |
Wood, S. A., Biessy, L., Latchford, J. L., Zaiko, A., von Ammon, U., Audrezet, F., Cristescu, M. E., and Pochon, X. (2020). Release and degradation of environmental DNA and RNA in a marine system. The Science of the Total Environment 704, 135314.
| Release and degradation of environmental DNA and RNA in a marine system.Crossref | GoogleScholarGoogle Scholar | 31780169PubMed |
Zencich, S. J., Froend, R. H., Turner, J. V., and Gailitis, V. (2002). Influence of groundwater depth on the seasonal sources of water accessed by Banksia tree species on a shallow, sandy coastal aquifer. Oecologia 131, 8–19.
| Influence of groundwater depth on the seasonal sources of water accessed by Banksia tree species on a shallow, sandy coastal aquifer.Crossref | GoogleScholarGoogle Scholar | 28547514PubMed |
Zhu, B. (2006). Degradation of plasmid and plant DNA in water microcosms monitored by natural transformation and real-time polymerase chain reaction (PCR). Water Research 40, 3231–3238.
| Degradation of plasmid and plant DNA in water microcosms monitored by natural transformation and real-time polymerase chain reaction (PCR).Crossref | GoogleScholarGoogle Scholar | 16945402PubMed |
Zinger, L., Chave, J., Coissac, E., Iribar, A., Louisanna, E., Manzi, S., Schilling, V., Schimann, H., Sommeria-Klein, G., and Taberlet, P. (2016). Extracellular DNA extraction is a fast, cheap and reliable alternative for multi-taxa surveys based on soil DNA. Soil Biology & Biochemistry 96, 16–19.
| Extracellular DNA extraction is a fast, cheap and reliable alternative for multi-taxa surveys based on soil DNA.Crossref | GoogleScholarGoogle Scholar |
Zinger, L., Bonin, A., Alsos, I. G., Bálint, M., Bik, H., Boyer, F., Chariton, A. A., Creer, S., Coissac, E., Deagle, B. E., and De Barba, M. (2019). DNA metabarcoding: need for robust experimental designs to draw sound ecological conclusions. Molecular Ecology 28, 1857–1862.
| DNA metabarcoding: need for robust experimental designs to draw sound ecological conclusions.Crossref | GoogleScholarGoogle Scholar | 31033079PubMed |


