A muddy time capsule: using sediment environmental DNA for the long-term monitoring of coastal vegetated ecosystems
N. R. Foster A D , B. M. Gillanders A , A. R. Jones A , J. M. Young B and M. Waycott A C
A C
A School of Biological Sciences, University of Adelaide, Adelaide, SA 5005, Australia.
B College of Science and Engineering, Flinders University, Bedford Park, SA 5042, Australia.
C State Herbarium of South Australia, Botanic Gardens and State Herbarium, Department of Environment and Water (DEW), Adelaide, SA 5001, Australia.
D Corresponding author. Email: nicole.foster@adelaide.edu.au
Marine and Freshwater Research 71(8) 869-876 https://doi.org/10.1071/MF19175
Submitted: 15 May 2019 Accepted: 23 January 2020 Published: 16 March 2020
Journal Compilation © CSIRO 2020 Open Access CC BY-NC-ND
Abstract
Seagrass, saltmarsh and mangrove habitats are declining around the world as anthropogenic activity and climate change intensify. To be able to effectively restore and maintain healthy coastal-vegetation communities, we must understand how and why they have changed in the past. Identifying shifts in vegetation communities, and the environmental or human drivers of these, can inform successful management and restoration strategies. Unfortunately, long-term data (i.e. decades to hundreds of years) on coastal vegetated ecosystems that can discern community-level changes are mostly non-existent in the scientific record. We propose implementing DNA extracted from coastal sediments to provide an alternative approach to long-term ecological reconstruction for coastal vegetated ecosystems. This type of DNA is called ‘environmental DNA’ and has previously been used to generate long-term datasets for other vegetated systems but has not yet been applied to vegetation change in coastal settings. In this overview, we explore the idea of using sediment eDNA as a long-term monitoring tool for seagrass, saltmarsh and mangrove communities. We see real potential in this approach for reconstructing long-term ecological histories of coastal vegetated ecosystems, and advocate that further research be undertaken to develop appropriate methods for its use.
Additional keywords: climate change, eDNA, mangrove, saltmarsh, seagrass.
Introduction
Seagrasses, saltmarshes and mangroves are plant communities that live along coastlines globally. Unfortunately, these environments are in decline worldwide, primarily as a result of human activities in coastal areas and, more recently, the effects of climate change (Alongi 2008; Gedan et al. 2009; Waycott et al. 2009). Protection and conservation of these coastal vegetation communities are extremely important because of the many valuable ecosystem services they provide (Costanza et al. 2014). Such services include providing breeding and migration habitats for marine life and birds, nutrient cycling and carbon storage, stabilising coastlines against erosion and sea-level rise, and supporting a range of industries, including fisheries and tourism (Barbier et al. 2011; Mcleod et al. 2011; Rogers et al. 2019). The specific ecosystem services provided by seagrasses, saltmarshes and mangroves are dependent on the distinctive species composition, morphology and habitat range of each community. Thus, coastal plant communities are highly diverse in their ecosystem service provision and all are essential to the survival of coastal ecosystems as a whole.
Our ability to protect coastal vegetated environments and prevent further decline rests on our understanding of how these communities have changed in the past in response to human stressors and environmental change. Long-term monitoring data that are high in resolution and span informative timescales (i.e. decades to hundreds of years), provide the ability to contextualise present conditions and predict future scenarios (Costanza et al. 2007). Change data that predate colonisation and industrialisation can help us understand ecological baselines and natural variability for coastal vegetated ecosystems, leading to an improved assessment of the impact of intensified human activities, the resilience of the system and the dynamics and time frames associated with recovery from disturbance (Bálint et al. 2018). Long-term data can also improve our understanding of invasion events and biotic interactions, further informing conservation policies and management efforts, and increasing their chances of success (Bálint et al. 2018). These important long-term change data are becoming increasingly sought after and, yet, are largely lacking for many ecosystems, including coastal environments (Boero et al. 2015).
Environmental DNA (eDNA), which is the DNA retained in environmental samples such as soil and water, can be extracted from coastal sediment cores and analysed to provide a new approach for obtaining long-term data on seagrass, saltmarsh and mangrove communities. eDNA from these coastal plant groups is preserved in sediment layers in various forms, including both active and dormant tissues, seeds, pollen and detritus (Fahner et al. 2016). This eDNA can be extracted and identified by matching the sequences obtained from the samples to those in an existing reference database. Sediment cores can be dated using radioisotopes, providing a core chronology where each layer has an estimated date that can be related to the coastal plant identification provided by eDNA analysis. The notion of using environmental DNA from sediment cores to reconstruct past vegetated environments is a well documented approach for gathering long-term data (Thomsen and Willerslev 2015; Bálint et al. 2018), but has not yet been applied to coastal vegetation systems. In this paper, we outline an environmental DNA ‘toolkit’ that can be adopted for assessing change in coastal plant communities, using samples from sediment cores. We highlight the benefits of this proposed approach over other existing monitoring methods, identify the emerging technologies that can support the analyses, and discuss how the resulting datasets will benefit the conservation, restoration and management of valuable coastal vegetated ecosystems.
Long-term monitoring of coastal vegetated environments; existing methods and an eDNA approach
Existing methods to document long-term change
Current approaches to collecting long-term data on seagrass, saltmarsh and mangrove environments include remote sensing, field-based observations, recovery of fossils and indirect estimates of vegetation composition based on molecular proxies and stable isotopes (Powell and Steele 1995). Remote sensing and field observations make it possible to evaluate change over metres to kilometres spatially; however, multiple surveys over time are needed to generate a time-series of records for change detection, and continued observations can be costly and time consuming (Danovaro et al. 2016). Additional field data can be generated at a lower cost when they are re-purposed as in Foster et al. (2017), where they applied underwater photography initially taken for estimating sediment accumulation, to the presence/absence of seagrass communities in South Australia. Despite the usefulness of these approaches, field observations date back only to when humans began recording their observations, and remote-sensing data are available only since the implementation of satellite-mounted instruments, with global, high-resolution data being limited to the past four decades.
In contrast, direct observation of plant communities using fossils enables assessment over geological timescales, but in a very site specific (spatially limited) and typically fragmentary manner. Indirect observations of past coastal vegetation can be made through chemical analysis of coastal sediments (using lipid biomarkers and stable isotopes). These data provide broad inference of climate and ecosystem changes that can be evaluated over decades to thousands of years (Ellegaard et al. 2006; Johnson et al. 2007; Cohen et al. 2012). However, this approach provides limited information regarding community composition and taxonomic resolution, which is a significant limitation when attempting to reconstruct the ecological history of a coastal site (Duffy et al. 2019). Owing to either a temporal limitation, limited taxonomic resolution or a general lack of data, current methods for monitoring long-term change are difficult to apply to answer many of the questions we have around change in coastal vegetated environments.
Why an eDNA approach?
We propose that long-term data on the dynamics of coastal vegetated ecosystems can be improved by analysing environmental DNA buried in the sediment layers under these communities, to identify when and how plant communities have changed over time. These changes can be dated (with radioisotope approaches), which will allow the ecological history of the site to be reconstructed over periods of hundreds to thousands of years. This chronology of vegetation dynamics can then be related to other datasets, such as, for example, on management actions, human activities or environmental change. This approach will allow a longer-term view of coastal plant-community baselines and how they have historically responded to changes in their environment. Any correlations that can be identified between vegetation-community change and environmental change can provide evidence to predict the response of the ecosystem to future changes. A long-term dataset on vegetation-community dynamics will also allow a better understanding of the natural state of coastal vegetation ecosystems, acting as a potential baseline for conservation, restoration and management strategies (Fig. 1).
Several studies have already applied an eDNA reconstruction approach to terrestrial plant communities. Giguet-Covex et al. (2014) related plant dynamics to livestock farming over millennia, correlating farming practices to vegetation changes; Pansu et al. (2015) identified vegetation shifts from forest to pastoral plant assemblages, which were found to be due to human modification such as burning, logging and grazing. Additionally, Ficetola et al. (2018) found that plant-community change was rapid after the introduction of rabbits to a remote island. It is unclear why there has been a lack of eDNA studies in coastal environments, because the environmental properties of coastal sediments, such as high salt concentrations, protection from UV, low temperature and the absence of oxygen, are all known to favour DNA preservation (Giguet-Covex et al. 2019). However, there is uncertainty around using eDNA recovered from coastal sediments, owing to the influence of tidal flow. Additionally, seawater flushing (horizontal) and water seepage (vertical) through sediment layers may influence the reliability of reconstructed ecological chronologies. For example, there is the risk that seawater can seep vertically through layers of sediment, transporting fragments of DNA from one layer to the next, thus smearing the signal of a vegetation community across multiple sediment layers (i.e. time periods).
We acknowledge that movement of genetic material could potentially hinder the effective use of eDNA in reconstructing coastal vegetation dynamics over time. However, previous literature has pointed to eDNA being representative of localised vegetation in habitats with water movement. Shackleton et al. (2019) were able to successfully use eDNA to reconstruct spatial patterns in plant communities within floodplains and Jeunen et al. (2019) found that eDNA surveys detected taxa that were spatially discrete in a coastal setting, noting that localised species generated local signals. Additionally, eDNA extracted from the soil at a specific location has been proven to reliably reflect aboveground plant species in a terrestrial environment (Yoccoz et al. 2012). For long-term data, eDNA complements and confirms results of pollen analysis dating back to the early Holocene in a lake system (Paus et al. 2015) as well as macrofossils and pollen in a late glacial lake (Parducci et al. 2019). Because of this evidence, we feel that eDNA analysis should certainly be trialled in coastal vegetated environments because the potential benefits to management would be considerable (See section ‘Using eDNA data to improve outcomes for coastal vegetated ecosystems’).
Common methods applied to environmental DNA analysis
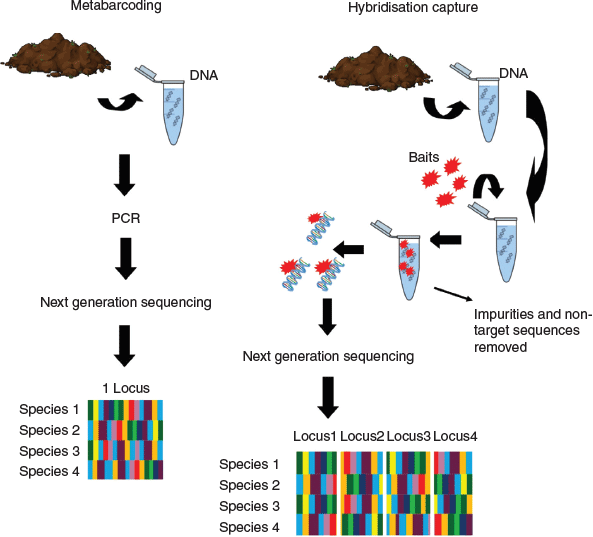
Metabarcoding of environmental DNA from sediments is a common approach to generating data on plant communities in soil (Zinger et al. 2019). DNA metabarcoding allows the identification of multiple species from a single environmental sample (Taberlet et al. 2012). This method was made possible because of the advances in DNA sequencing technologies and was further refined through the use of next-generation sequencing (NGS) that can read DNA from multiple samples in parallel. When using this approach, DNA is extracted from a sediment sample and then either one or multiple universal barcode regions are selected and amplified by primers via polymerase chain reaction (PCR) and then sequenced using NGS. With metabarcoding, the DNA barcode locus used must have gene regions that are short enough to allow adequate amplification from degraded DNA fragments, which are generally found in environmental samples. These fragments are shorter than DNA recovered from fresh plant tissue, that is, live plants (Epp et al. 2012). This means that the choice of barcode is essential so as to recover all the taxa present in the sediment sample. A commonly used universal locus for detecting plant DNA in sediments is the P6 loop of the trnL intron (Taberlet et al. 2007). This is a short barcode, which is, therefore, more likely to be detected in sediments than is a longer barcode region. However, although a shorter barcode has a greater likelihood of recovering plant species present in the sediment, the reliance on this single short barcode means that taxonomic resolution will likely be only to family or order level and, in rare instances, genus level. Therefore, depending on the questions to be answered, that is, whether broader or finer taxonomic resolution is required, will determine whether metabarcoding is the appropriate choice, or whether alternative methods are required.
Recent eDNA-method developments
Hybridisation capture is a new approach that can be applied to analysing eDNA from sediment cores, which can potentially improve on metabarcoding. Instead of applying a single universal locus per PCR, many loci are targeted via the design of custom, short RNA probes called ‘baits’ (Fig. 2). This reduces the reliance on a single PCR step which can lead to biases related to primers preferentially binding to particular species or the absence of primer binding sites. To design baits, target species genomes are selected, and these are then used to generate very specific target loci (Lemmon et al. 2012). If multiple baits are designed, and they capture at the family or order level (Hart et al. 2016), then multiple species can be targeted using the same bait set. Recently, a bait set was designed by Waycott et al. (M. Waycott, E. Biffin, K. Van-Dijk, unpubl data) to capture across all flowering plants in a single capture, that is, the baits are not designed to be species specific but are designed to capture multiple species. This not only decreases costs but also increases the chance of detecting all taxa present in a sediment sample. Once DNA has been extracted from the sediment, the hybridisation capture protocol includes assigning unique identification codes to samples, which is a series of unique bases attached to the ends of the reads. This means that multiple samples can be pooled and they can be separated in the analysis step, this is ideal because it further reduces costs of this approach. Baits are then added to these pooled samples and the bait–DNA combination is removed from solution by using magnetic beads, thus leaving behind only unwanted sequences and impurities, which can then be discarded. The targeted sequences that have been removed are then amplified via PCR and sequenced using NGS. Because of the degradation of DNA in sediments, single target loci can be difficult or impossible to recover for every species. By targeting many loci instead of one, there is a greater probability of detecting the eDNA left behind by all individual coastal plants present in the sediment. The discriminatory ability of multiple loci also allows greater taxonomic resolution to be achieved when identifying eDNA sequences recovered from sediments. Overall, we suggest that hybridisation capture could provide a highly effective tool for analysing eDNA from coastal sediments, where it is important to document all taxa present.
|
Is hybridisation capture feasible and what information can we obtain?
A hybridisation-capture approach for genomic analysis of multiple species in a single sample has already shown promising results and prompted consideration of how this information can fill knowledge gaps in other disciplines. Recent work by Foster et al. (N. R. Foster, J. M. Young, K. Van-Dijk, E. Biffin, A. R. Jones, B. M. Gillanders, M. Waycott, unpubl. data) has demonstrated that hybridisation capture can consistently identify individual coastal plant species in a mock sample of up to 10 different species. This was then tested on modern sediments, that is, only ~20 years before present, where the results confirmed that a hybridisation-capture approach is able to recover species-level information from eDNA in sediments. Further testing will need to be undertaken to determine the temporal limitations of this approach; however, the current results have suggested that this method can recover high-resolution information for individual plant species within an environmental sample. Such information can be applied to determining sources of organic carbon in coastal plant environments as in Reef et al. (2017). Extrapolating these data to long-term trends will provide insight into which vegetation communities were responsible for the historic sequestration of stored organic carbon in coastal sediments (Geraldi et al. 2019), which can then inform the way these environments are managed. The species-specific genetic data uncovered with this approach can also be applied to population-level variability within species because there is a higher likelihood of detecting within-species variability because multiple genes are targeted. Within-species variability can provide information on recruitment and effective population size, which are useful metrics for understanding vegetation community dynamics. In addition, population-genetic applications are widely used in management of natural systems (Adams et al. 2019). Ongoing advances in the application of eDNA, such as the suggested hybridisation capture or even genome skimming, that avoid the inherent biases of PCR-based methods, are enabling more reliable eDNA-based population genetic data to be generated. Work is ongoing to overcome remaining challenges, including testing to remove confounding factors (e.g. degraded DNA leading to misidentification of sequence variation within a species; Adams et al. 2019). Evidently, a hybridisation-capture approach is not only feasible but can supplement data in other environmental research areas that are otherwise difficult to obtain.
Additional considerations when applying an eDNA approach to coastal systems
The application of eDNA for monitoring coastal vegetation change is certainly feasible; however, a major limiting factor of this approach is the availability of reference sequences for coastal plant communities. A taxonomically verified and high-quality reference sequence database is required to identify DNA sequences extracted from coastal sediment samples. Although unknown sequences and assignment of sequences to a higher order can still be useful, sequence data are far more useful when they can be accurately classified to a species or genus (Reef et al. 2017; Cristescu and Hebert 2018; Dormontt et al. 2018).
So as to construct a reference database, species of interest need to be properly vouchered with correct taxonomic identification and deposited in Herbaria (Dormontt et al. 2018). A lack of vouchered specimens and their associated DNA sequences significantly impairs progress (Thomsen and Willerslev 2015) and reduces our ability to identify sequences, which affects the accuracy and utility of the results (Jørgensen et al. 2012). False positives may be mistakenly generated where coastal plant species are believed to have been present because they have a partial match to a sequence in an incomplete reference database. Conversely, a coastal plant sequence may be generated but not have a match in the reference database, leading to it be mistakenly discarded. While there is software available to decrease the likelihood of a false classification, a complete reference library will still greatly improve the accurate assignment of eDNA sequences to known species.
Reference libraries for plant communities are being constructed, such as the database on arctic flora initiated by Sønstebø et al. (2010), which was then expanded by (Boessenkool et al. 2014). However, coastal plant reference databases are yet to be constructed or made widely available (Duffy et al. 2019); therefore, this is an area of research that needs to be addressed before implementing eDNA as a management tool. Additionally, reference sequences must also be of the same loci as chosen for amplification of the DNA from sediments, that is, either a universal locus in the case of metabarcoding or multiple loci in the case of hybridisation capture. Fortunately, if the latter is chosen, it is a faster and more cost-effective approach and will generate significantly more reference data because of the fact that samples can be pooled, and many loci are targeted instead of one.
Using eDNA data to improve outcomes for coastal vegetated ecosystems
Effective management and conservation of coastal vegetated ecosystems requires a long-term outlook that is difficult to achieve with existing monitoring methods, but critical for understanding ecosystem dynamics (Duffy et al. 2019; McAfee et al. 2019). For example, Watson et al. (2011) used pollen and isotope evidence from dated cores to establish a long-term ecological history for Elkhorn Slough tidal marsh in California. Reconstruction of marsh community composition over a period of 5000 years considerably altered the perspective within which recent changes (in the past few decades) have been interpreted (Jackson et al. 2001). Using long-term datasets to establish ecological baselines, and understand the bounds of natural variability, greatly improves our ability to predict the likely ecosystem response to future changes, on the basis of its response to past change (Fordham et al. 2016; Nogués-Bravo et al. 2018), with higher temporal and taxonomic resolution datasets providing greater gains (Duffy et al. 2019; García 2019). The proposed eDNA approach for reconstructing coastal vegetation ecosystem dynamics will allow generation of well resolved taxonomic data time series (i.e. species presence and community composition) over hundreds to thousands of years at approximately decadal temporal resolution for specific sample locations.
The foundational information that can be obtained from such long time series include the following: identification of historically stable vegetation communities at specific locations, which may be used as a target for native vegetation restoration; identification and timing of invasive species introductions, and the subsequent responses of native vegetation communities to invasions; detection of rare, uncommon or cryptic species that are difficult to monitor by using other methods; and biotic responses of coastal vegetation communities to stressors such as coastal habitat modification and sea-level rise. As such, long-term eDNA-based ecological reconstructions of coastal vegetation can provide scientific understanding to underpin policy and management decisions. Below we have outlined a few key examples of potential applications of these data.
Sediment core samples from multiple locations could be analysed to generate data on the spatial and temporal patterns of coastal vegetation species’ range shifts, local extirpations, evolutionary adaptations and extinctions in response to climate change over periods of potentially thousands of years (depending on the sediment age profile). The same datasets could also be used to assess the responses of different taxa to human activities, including understanding how cumulative impacts may push coastal vegetation communities towards tipping points and result in significantly altered species compositions. It may also be possible to use these well resolved eDNA-based time series to assess the success or failure of previous management actions (e.g. attempts at restoration) and inform adaptive management approaches; to better understand the time frames required to restore ecosystems to their baseline state; and ultimately to assess whether the conditions required for restoration of the stable baseline community are realistically achievable under current and future climatic conditions (Breed et al. 2016).
In addition to the above practical applications, eDNA-based reconstruction of coastal vegetation environmental histories can make a difference for public engagement and the public perception of coastal wetland ecosystems. As noted by McAfee et al. (2019), using ecological reconstructions to create a historical context for proposed restoration projects can generate confidence by clearly demonstrating tangible targets, and may also positively affect the intrinsic value that stakeholders attribute to an ecosystem.
Conclusions
Ongoing climate change and negative impacts from human activities in the coastal zone threaten the survival of coastal vegetation communities and the enormous ecosystem services they provide. We need new approaches that can generate accurate, long-term data on the presence and species composition of these ecosystems over long time periods (up to thousands of years), because this provides vital information for conservation plans, future predictions, ecosystem processes and successful management policies. Environmental DNA is an emerging toolkit that is already being applied to monitoring of vegetation and aquatic species in other environmental settings and has great potential to be implemented in coastal vegetated environments. We advocate for novel methods such as hybridisation capture to continue to be trialled on dated sediments sampled from coastal vegetated environments, because this will only improve the quantity and quality of the taxonomic information generated. There is great potential for this proposed novel approach to generate previously unobtainable ecological histories for coastal vegetated ecosystems. Armed with such datasets, we can better understand their long-term dynamics, engage stakeholders, inform successful management and ultimately achieve positive outcomes for these valuable ecosystems into the future.
Conflicts of interest
N. R. Foster and M. Waycott are Guest Editors of the Translating Seagrass Science into Action issue and B. M. Gillanders is an Associate Editor with Marine and Freshwater Research. Despite these relationships, they took no part in the review and acceptance of this or any other manuscript in this issue that they authored. The authors declare that they have no further conflicts of interest.
Declaration of funding
N. R. Foster was supported by a University of Adelaide student Ph.D. scholarship. Funding for A. R. Jones was provided by a grant from the Goyder Institute for Water Research, with co-funding received from SA Water and the University of Adelaide’s Environment Institute. Funding for this work was also provided by the Hermon Slade Foundation (HSF 18/2).
Acknowledgements
We thank the reviewers of this manuscript for their helpful comments and insights.
References
Adams, C. I. M., Knapp, M., Gemmell, N. J., Jeunen, G. J., Bunce, M., Lamare, M. D., and Taylor, H. R. (2019). Beyond biodiversity: can environmental DNA (eDNA) cut it as a population genetics tool? Genes 10, 192.| Beyond biodiversity: can environmental DNA (eDNA) cut it as a population genetics tool?Crossref | GoogleScholarGoogle Scholar |
Alongi, D. M. (2008). Mangrove forests: resilience, protection from tsunamis, and responses to global climate change. Estuarine, Coastal and Shelf Science 76, 1–13.
| Mangrove forests: resilience, protection from tsunamis, and responses to global climate change.Crossref | GoogleScholarGoogle Scholar |
Bálint, M., Pfenninger, M., Grossart, H. P., Taberlet, P., Vellend, M., Leibold, M. A., Englund, G., and Bowler, D. (2018). Environmental DNA time series in ecology. Trends in Ecology & Evolution 33, 945–957.
| Environmental DNA time series in ecology.Crossref | GoogleScholarGoogle Scholar |
Barbier, E. B., Hacker, S. D., Kennedy, C., Koch, E. W., Stier, A. C., and Silliman, B. R. (2011). The value of estuarine and coastal ecosystem services. Ecological Monographs 81, 169–193.
| The value of estuarine and coastal ecosystem services.Crossref | GoogleScholarGoogle Scholar |
Boero, F., Kraberg, A. C., Krause, G., and Wiltshire, K. H. (2015). Time is an affliction: why ecology cannot be as predictive as physics and why it needs time series. Journal of Sea Research 101, 12–18.
| Time is an affliction: why ecology cannot be as predictive as physics and why it needs time series.Crossref | GoogleScholarGoogle Scholar |
Boessenkool, S., McGlynn, G., Epp, L. S., Taylor, D., Pimentel, M., Gizaw, A., Nemomissa, S., Brochmann, C., and Popp, M. (2014). Use of ancient sedimentary DNA as a novel conservation tool for high-altitude tropical biodiversity. Conservation Biology 28, 446–455.
| Use of ancient sedimentary DNA as a novel conservation tool for high-altitude tropical biodiversity.Crossref | GoogleScholarGoogle Scholar | 24372820PubMed |
Breed, M. F., Lowe, A. J., and Mortimer, P. E. (2016). Restoration: ‘garden of Eden’ unrealistic. Nature 533, 469.
| Restoration: ‘garden of Eden’ unrealistic.Crossref | GoogleScholarGoogle Scholar | 27225110PubMed |
Cohen, M. C. L., Pessenda, L. C. R., Behling, H., de Fátima Rossetti, D., França, M. C., Guimarães, J. T. F., Friaes, Y., and Smith, C. B. (2012). Holocene palaeoenvironmental history of the Amazonian mangrove belt. Quaternary Science Reviews 55, 50–58.
| Holocene palaeoenvironmental history of the Amazonian mangrove belt.Crossref | GoogleScholarGoogle Scholar |
Costanza, R., Graumlich, L., Steffen, W., Crumley, C., Dearing, J., Hibbard, K., Leemans, R., Redman, C., and Schimel, D. (2007). Sustainability or collapse: what can we learn from integrating the history of humans and the rest of nature? Ambio 36, 522–527.
| Sustainability or collapse: what can we learn from integrating the history of humans and the rest of nature?Crossref | GoogleScholarGoogle Scholar | 18074887PubMed |
Costanza, R., de Groot, R., Sutton, P., van der Ploeg, S., Anderson, S. J., Kubiszewski, I., Farber, S., and Turner, R. K. (2014). Changes in the global value of ecosystem services. Global Environmental Change 26, 152–158.
| Changes in the global value of ecosystem services.Crossref | GoogleScholarGoogle Scholar |
Cristescu, M. E., and Hebert, P. D. (2018). Uses and misuses of environmental DNA in biodiversity science and conservation. Annual Review of Ecology, Evolution, and Systematics 49, 209–230.
| Uses and misuses of environmental DNA in biodiversity science and conservation.Crossref | GoogleScholarGoogle Scholar |
Danovaro, R., Carugati, L., Berzano, M., Cahill, A. E., Carvalho, S., Chenuil, A., Corinaldesi, C., Cristina, S., David, R., Dell’Anno, A., Dzhembekova, N., Garcés, E., Gasol, J. M., Goela, P., Féral, J.-P., Ferrera, I., Forster, R. M., Kurekin, A. A., Rastelli, E., Marinova, V., Miller, P. I., Moncheva, S., Newton, A., Pearman, J. K., Pitois, S. G., Reñé, A., Rodríguez-Ezpeleta, N., Saggiomo, V., Simis, S. G. H., Stefanova, K., Wilson, C., Lo Martire, M., Greco, S., Cochrane, S. K. J., Mangoni, O., and Borja, A. (2016). Implementing and innovating marine monitoring approaches for assessing marine environmental status. Frontiers in Marine Science 3, 213.
| Implementing and innovating marine monitoring approaches for assessing marine environmental status.Crossref | GoogleScholarGoogle Scholar |
Dormontt, E. E., Van Dijk, K.-j., Bell, K. L., Biffin, E., Breed, M. F., Byrne, M., Caddy-Retalic, S., Encinas-Viso, F., Neville, P., and Shapcott, A. (2018). Advancing DNA metabarcoding applications for plants requires systematic analysis of herbarium collections: an Australian perspective. Frontiers in Ecology and Evolution 6, 134.
| Advancing DNA metabarcoding applications for plants requires systematic analysis of herbarium collections: an Australian perspective.Crossref | GoogleScholarGoogle Scholar |
Duffy, J. E., Benedetti-Cecchi, L., Trinanes, J., Muller-Karger, F. E., Ambo-Rappe, R., Boström, C., Buschmann, A. H., Byrnes, J., Coles, R. G., Creed, J., Cullen-Unsworth, L. C., Diaz-Pulido, G., Duarte, C. M., Edgar, G. J., Fortes, M., Goni, G., Hu, C., Huang, X., Hurd, C. L., Johnson, C., Konar, B., Krause-Jensen, D., Krumhansl, K., Macreadie, P., Marsh, H., McKenzie, L. J., Mieszkowska, N., Miloslavich, P., Montes, E., Nakaoka, M., Norderhaug, K. M., Norlund, L. M., Orth, R. J., Prathep, A., Putman, N. F., Samper-Villarreal, J., Serrao, E. A., Short, F., Pinto, I. S., Steinberg, P., Stuart-Smith, R., Unsworth, R. K. F., van Keulen, M., van Tussenbroek, B. I., Wang, M., Waycott, M., Weatherdon, L. V., Wernberg, T., and Yaakub, S. M. (2019). Toward a coordinated global observing system for seagrasses and marine macroalgae. Frontiers in Marine Science 6, 317.
| Toward a coordinated global observing system for seagrasses and marine macroalgae.Crossref | GoogleScholarGoogle Scholar |
Ellegaard, M., Clarke, A. L., Reuss, N., Drew, S., Weckström, K., Juggins, S., Anderson, N. J., and Conley, D. J. (2006). Multi-proxy evidence of long-term changes in ecosystem structure in a Danish marine estuary, linked to increased nutrient loading. Estuarine, Coastal and Shelf Science 68, 567–578.
| Multi-proxy evidence of long-term changes in ecosystem structure in a Danish marine estuary, linked to increased nutrient loading.Crossref | GoogleScholarGoogle Scholar |
Epp, L. S., Boessenkool, S., Bellemain, E. P., Haile, J., Esposito, A., Riaz, T., Erseus, C., Gusarov, V. I., Edwards, M. E., Johnsen, A., Stenoien, H. K., Hassel, K., Kauserud, H., Yoccoz, N. G., Brathen, K. A., Willerslev, E., Taberlet, P., Coissac, E., and Brochmann, C. (2012). New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Molecular Ecology 21, 1821–1833.
| New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems.Crossref | GoogleScholarGoogle Scholar | 22486821PubMed |
Fahner, N. A., Shokralla, S., Baird, D. J., and Hajibabaei, M. (2016). Large-scale monitoring of plants through environmental DNA metabarcoding of soil: recovery, resolution, and annotation of four DNA markers. PLoS One 11, e0157505.
| Large-scale monitoring of plants through environmental DNA metabarcoding of soil: recovery, resolution, and annotation of four DNA markers.Crossref | GoogleScholarGoogle Scholar | 27310720PubMed |
Ficetola, G. F., Poulenard, J., Sabatier, P., Messager, E., Gielly, L., Leloup, A., Etienne, D., Bakke, J., Malet, E., and Fanget, B. (2018). DNA from lake sediments reveals long-term ecosystem changes after a biological invasion. Science Advances 4, eaar4292.
| DNA from lake sediments reveals long-term ecosystem changes after a biological invasion.Crossref | GoogleScholarGoogle Scholar | 29750197PubMed |
Fordham, D. A., Akçakaya, H. R., Alroy, J., Saltré, F., Wigley, T. M. L., and Brook, B. W. (2016). Predicting and mitigating future biodiversity loss using long-term ecological proxies. Nature Climate Change 6, 909–916.
| Predicting and mitigating future biodiversity loss using long-term ecological proxies.Crossref | GoogleScholarGoogle Scholar |
Foster, N. R., Fotheringham, D. G., Brock, D. J., and Waycott, M. (2017). A resourceful and adaptable method to obtain data on the status of seagrass meadows. Aquatic Botany 141, 17–21.
| A resourceful and adaptable method to obtain data on the status of seagrass meadows.Crossref | GoogleScholarGoogle Scholar |
García, C. (2019). From ecological indicators to ecological functioning: integrative approaches to seize on ecological, climatic and socio-economic databases. Ecological Indicators 107, 105612.
| From ecological indicators to ecological functioning: integrative approaches to seize on ecological, climatic and socio-economic databases.Crossref | GoogleScholarGoogle Scholar |
Gedan, K. B., Silliman, B. R., and Bertness, M. D. (2009). Centuries of human-driven change in salt marsh ecosystems. Annual Review of Marine Science 1, 117–141.
| Centuries of human-driven change in salt marsh ecosystems.Crossref | GoogleScholarGoogle Scholar | 21141032PubMed |
Geraldi, N. R., Ortega, A., Serrano, O., Macreadie, P. I., Lovelock, C. E., Krause-Jensen, D., Kennedy, H., Lavery, P. S., Pace, M. L., Kaal, J., and Duarte, C. M. (2019). Fingerprinting blue carbon: rationale and tools to determine the source of organic carbon in marine depositional environments. Frontiers in Marine Science 6, 263.
| Fingerprinting blue carbon: rationale and tools to determine the source of organic carbon in marine depositional environments.Crossref | GoogleScholarGoogle Scholar |
Giguet-Covex, C., Pansu, J., Arnaud, F., Rey, P. J., Griggo, C., Gielly, L., Domaizon, I., Coissac, E., David, F., Choler, P., Poulenard, J., and Taberlet, P. (2014). Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nature Communications 5, 3211.
| Long livestock farming history and human landscape shaping revealed by lake sediment DNA.Crossref | GoogleScholarGoogle Scholar | 24487920PubMed |
Giguet-Covex, C., Ficetola, G. F., Walsh, K. J., Poulenard, J., Bajard, M., Fouinat, L., Sabatier, P., Gielly, L., Messager, E., Develle, A.-L., David, F., Taberlet, P., Brisset, E., Guiter, F., Sinet, R., and Arnaud, F. (2019). New insights on lake sediment DNA from the catchment: importance of taphonomic and analytical issues on the record quality. Scientific Reports 9, .
| New insights on lake sediment DNA from the catchment: importance of taphonomic and analytical issues on the record quality.Crossref | GoogleScholarGoogle Scholar | 31604959PubMed |
Hart, M. L., Forrest, L. L., Nicholls, J. A., and Kidner, C. A. (2016). Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 65, 1081–1092.
| Retrieval of hundreds of nuclear loci from herbarium specimens.Crossref | GoogleScholarGoogle Scholar |
Jackson, J. B., Kirby, M. X., Berger, W. H., Bjorndal, K. A., Botsford, L. W., Bourque, B. J., Bradbury, R. H., Cooke, R., Erlandson, J., and Estes, J. A. (2001). Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637.
| Historical overfishing and the recent collapse of coastal ecosystems.Crossref | GoogleScholarGoogle Scholar | 11474098PubMed |
Jeunen, G. J., Knapp, M., Spencer, H. G., Lamare, M. D., Taylor, H. R., Stat, M., Bunce, M., and Gemmell, N. J. (2019). Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Molecular Ecology Resources 19, 426–438.
| Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement.Crossref | GoogleScholarGoogle Scholar | 30576077PubMed |
Johnson, B. J., Moore, K. A., Lehmann, C., Bohlen, C., and Brown, T. A. (2007). Middle to late Holocene fluctuations of C3 and C4 vegetation in a Northern New England Salt Marsh, Sprague Marsh, Phippsburg Maine. Organic Geochemistry 38, 394–403.
| Middle to late Holocene fluctuations of C3 and C4 vegetation in a Northern New England Salt Marsh, Sprague Marsh, Phippsburg Maine.Crossref | GoogleScholarGoogle Scholar |
Jørgensen, T., Kjaer, K. H., Haile, J., Rasmussen, M., Boessenkool, S., Andersen, K., Coissac, E., Taberlet, P., Brochmann, C., Orlando, L., Gilbert, M. T., and Willerslev, E. (2012). Islands in the ice: detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA meta-barcoding. Molecular Ecology 21, 1980–1988.
| Islands in the ice: detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA meta-barcoding.Crossref | GoogleScholarGoogle Scholar | 21951625PubMed |
Lemmon, A. R., Emme, S. A., and Lemmon, E. M. (2012). Anchored hybrid enrichment for massively high-throughput phylogenomics. Systematic Biology 61, 727–744.
| Anchored hybrid enrichment for massively high-throughput phylogenomics.Crossref | GoogleScholarGoogle Scholar | 22605266PubMed |
McAfee, D., Alleway, H. K., and Connell, S. D. (2019). Environmental solutions sparked by environmental history. Conservation Biology , .
| Environmental solutions sparked by environmental history.Crossref | GoogleScholarGoogle Scholar | 31385623PubMed |
Mcleod, E., Chmura, G. L., Bouillon, S., Salm, R., Björk, M., Duarte, C. M., Lovelock, C. E., Schlesinger, W. H., and Silliman, B. R. (2011). A blueprint for blue carbon: toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2. Frontiers in Ecology and the Environment 9, 552–560.
| A blueprint for blue carbon: toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2.Crossref | GoogleScholarGoogle Scholar |
Nogués-Bravo, D., Rodriguez-Sanchez, F., Orsini, L., de Boer, E., Jansson, R., Morlon, H., Fordham, D. A., and Jackson, S. T. (2018). Cracking the code of biodiversity responses to past climate change. Trends in Ecology & Evolution 33, 765–776.
| Cracking the code of biodiversity responses to past climate change.Crossref | GoogleScholarGoogle Scholar |
Pansu, J., Giguet-Covex, C., Ficetola, G. F., Gielly, L., Boyer, F., Zinger, L., Arnaud, F., Poulenard, J., Taberlet, P., and Choler, P. (2015). Reconstructing long-term human impacts on plant communities: an ecological approach based on lake sediment DNA. Molecular Ecology 24, 1485–1498.
| Reconstructing long-term human impacts on plant communities: an ecological approach based on lake sediment DNA.Crossref | GoogleScholarGoogle Scholar | 25735209PubMed |
Parducci, L., Alsos, I. G., Unneberg, P., Pedersen, M. W., Han, L., Lammers, Y., Salonen, J. S., Väliranta, M. M., Slotte, T., and Wohlfarth, B. (2019). Shotgun environmental DNA, pollen, and macrofossil analysis of late glacial lake sediments from southern Sweden. Frontiers in Ecology and Evolution 7, .
| Shotgun environmental DNA, pollen, and macrofossil analysis of late glacial lake sediments from southern Sweden.Crossref | GoogleScholarGoogle Scholar |
Paus, A., Boessenkool, S., Brochmann, C., Epp, L. S., Fabel, D., Haflidason, H., and Linge, H. (2015). Lake Store Finnsjøen: a key for understanding Lateglacial/early Holocene vegetation and ice sheet dynamics in the central Scandes Mountains. Quaternary Science Reviews 121, 36–51.
| Lake Store Finnsjøen: a key for understanding Lateglacial/early Holocene vegetation and ice sheet dynamics in the central Scandes Mountains.Crossref | GoogleScholarGoogle Scholar |
Powell, T. M., and Steele, J. H. (1995). ‘Ecological Time Series.’ (Chapman & Hall: New York, NY, USA.)
Reef, R., Atwood, T. B., Samper-Villarreal, J., Adame, M. F., Sampayo, E. M., and Lovelock, C. E. (2017). Using eDNA to determine the source of organic carbon in seagrass meadows. Limnology and Oceanography 62, 1254–1265.
| Using eDNA to determine the source of organic carbon in seagrass meadows.Crossref | GoogleScholarGoogle Scholar |
Rogers, K., Saintilan, N., Mazumder, D., and Kelleway, J. J. (2019). Mangrove dynamics and blue carbon sequestration. Biological Letters 15, 20180471.
| Mangrove dynamics and blue carbon sequestration.Crossref | GoogleScholarGoogle Scholar |
Shackleton, M. E., Rees, G. N., Watson, G., Campbell, C., and Nielsen, D. (2019). Environmental DNA reveals landscape mosaic of wetland plant communities. Global Ecology and Conservation 19, e00689.
| Environmental DNA reveals landscape mosaic of wetland plant communities.Crossref | GoogleScholarGoogle Scholar |
Sønstebø, J., Gielly, L., Brysting, A., Elven, R., Edwards, M., Haile, J., Willerslev, E., Coissac, E., Rioux, D., and Sannier, J. (2010). Using next‐generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Molecular Ecology Resources 10, 1009–1018.
| Using next‐generation sequencing for molecular reconstruction of past Arctic vegetation and climate.Crossref | GoogleScholarGoogle Scholar | 21565110PubMed |
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., Vermat, T., Corthier, G., Brochmann, C., and Willerslev, E. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Research 35, e14.
| Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding.Crossref | GoogleScholarGoogle Scholar | 17169982PubMed |
Taberlet, P., Coissac, E., Hajibabaei, M., and Rieseberg, L. H. (2012). Environmental DNA. Molecular Ecology 21, 1789–1793.
| Environmental DNA.Crossref | GoogleScholarGoogle Scholar | 22486819PubMed |
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA: an emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183, 4–18.
| Environmental DNA: an emerging tool in conservation for monitoring past and present biodiversity.Crossref | GoogleScholarGoogle Scholar |
Watson, E. B., Wasson, K., Pasternack, G. B., Woolfolk, A., Van Dyke, E., Gray, A. B., Pakenham, A., and Wheatcroft, R. A. (2011). Applications from paleoecology to environmental management and restoration in a dynamic coastal environment. Restoration Ecology 19, 765–775.
| Applications from paleoecology to environmental management and restoration in a dynamic coastal environment.Crossref | GoogleScholarGoogle Scholar |
Waycott, M., Duarte, C. M., Carruthers, T. J., Orth, R. J., Dennison, W. C., Olyarnik, S., Calladine, A., Fourqurean, J. W., Heck, K. L., and Hughes, A. R. (2009). Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proceedings of the National Academy of Sciences of the United States of America 106, 12377–12381.
| Accelerating loss of seagrasses across the globe threatens coastal ecosystems.Crossref | GoogleScholarGoogle Scholar | 19587236PubMed |
Yoccoz, N. G., Brathen, K. A., Gielly, L., Haile, J., Edwards, M. E., Goslar, T., Von Stedingk, H., Brysting, A. K., Coissac, E., Pompanon, F., Sonstebo, J. H., Miquel, C., Valentini, A., De Bello, F., Chave, J., Thuiller, W., Wincker, P., Cruaud, C., Gavory, F., Rasmussen, M., Gilbert, M. T., Orlando, L., Brochmann, C., Willerslev, E., and Taberlet, P. (2012). DNA from soil mirrors plant taxonomic and growth form diversity. Molecular Ecology 21, 3647–3655.
| DNA from soil mirrors plant taxonomic and growth form diversity.Crossref | GoogleScholarGoogle Scholar | 22507540PubMed |
Zinger, L., Bonin, A., Alsos, I. G., Balint, M., Bik, H., Boyer, F., Chariton, A. A., Creer, S., Coissac, E., Deagle, B. E., De Barba, M., Dickie, I. A., Dumbrell, A. J., Ficetola, G. F., Fierer, N., Fumagalli, L., Gilbert, M. T. P., Jarman, S., Jumpponen, A., Kauserud, H., Orlando, L., Pansu, J., Pawlowski, J., Tedersoo, L., Thomsen, P. F., Willerslev, E., and Taberlet, P. (2019). DNA metabarcoding: need for robust experimental designs to draw sound ecological conclusions. Molecular Ecology 28, 1857–1862.
| DNA metabarcoding: need for robust experimental designs to draw sound ecological conclusions.Crossref | GoogleScholarGoogle Scholar | 31033079PubMed |



