Characterisation of nematode larvae found in a vulnerable native Australian fish, the southern pygmy perch, Nannoperca australis Günther
Shokoofeh Shamsi A * , Luke Pearce B and Xiaocheng Zhu C D
A * , Luke Pearce B and Xiaocheng Zhu C D
A Gulbali Institute and School of Agricultural, Environmental and Veterinary Sciences, Charles Sturt University, Wagga Wagga, NSW 2678, Australia.
B NSW Department of Primary Industries, Fisheries, Habitat and Threatened Species Unit, Freshwater Environment Branch, Albury, NSW 2640, Australia.
C School of Agricultural, Environmental and Veterinary Sciences, Charles Sturt University, Wagga Wagga, NSW 2678, Australia.
D NSW Department of Primary Industries, Wagga Wagga Agricultural Institute, Wagga Wagga, NSW 2650, Australia.
Marine and Freshwater Research 74(12) 1095-1101 https://doi.org/10.1071/MF23095
Submitted: 18 May 2023 Accepted: 30 June 2023 Published: 27 July 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: The southern pygmy perch (Nannoperca australis) is an endemic freshwater fish in Australia that is facing population decline and is listed as endangered or vulnerable in several states.
Aims: The aim of this study was to investigate the presence of parasites in the southern pygmy perch population and provide insights into their effect on the health and conservation of the species.
Methods: In total, 81 southern pygmy perch specimens were examined for parasite infections, followed by characterisation of the parasites.
Key results: The postmortem examination of the fish specimens did not show any visible parasites. However, through the incubation method, nematode larvae were discovered in 14 fish (mean intensity 1.6, mean abundance 0.28).
Conclusions: This study represents the first report of nematode larvae belonging to the genus Spiroxys in Australia, specifically in the southern pygmy perch. These findings highlighted the presence of parasite infections in the endangered southern pygmy perch and underscored the importance of conducting further research on parasites and their potential effect on the health and conservation of this species.
Implications: The discovery of nematode larvae in the southern pygmy perch raises concerns about the potential effects of parasites on the population.
Keywords: detection method, endangered species, Gnathostomatidae, life cycle, native fish, Nematoda, parasites, wildlife.
Introduction
Southern pygmy perch (Nannoperca australis) is a small endemic freshwater fish species found in Australia (Allen 1989). It exhibits a maximum length of up to 85 mm, although it rarely exceeds 65 mm (FishBase, ver. 4.0, see https://www.fishbase.org.au/v4/summary/14920). Southern pygmy perch populations are primarily distributed in the south-eastern region of Australia, occupying various well-vegetated slow-flowing aquatic environments, such as gently flowing streams, lakes, billabongs, drains, dams, swamps, ephemeral creeks and wetlands. Their diet comprises a range of small aquatic crustaceans, insects and their larvae (Annonymous 2012).
In recent years, the southern pygmy perch has experienced significant population declines, particularly in New South Wales (Bray and Thompson 2022). These declines have resulted in severe fragmentation and range reductions, primarily within the Murray–Darling Basin. Consequently, the species has been classified as endangered, protected or vulnerable in several states because of various threats (Pearce et al. 2019). These threats include habitat degradation resulting from the loss of riparian and instream vegetation, increased sedimentation, poor water quality, river regulation reducing permanent floodplain habitats and dispersal opportunities, drought, flooding, predation and competition from introduced fish species such as redfin, brown trout, rainbow trout, eastern gambusia and common carp.
Despite the southern pygmy perch’s threatened status and declining population, limited information is available regarding the infectious agents that may cause diseases in this fish species. To the best of our knowledge, there is currently no existing report or study specifically addressing the parasites that infect southern pygmy perch. Therefore, the objective of this study was to report the occurrence of parasite infections in southern pygmy perch collected during a mortality event in February 2019.
Understanding the parasitic infections in endangered species such as the southern pygmy perch is crucial for effective conservation and management strategies. Parasites can exert significant impacts on the health, survival and reproductive success of host populations (Habibi and Shamsi 2018; Shamsi et al. 2021a). Additionally, parasites can serve as bioindicators of ecosystem health and environmental quality (Mascarenhas et al. 2021; Shamsi et al. 2023). By investigating parasite infections in the southern pygmy perch, valuable insights can be gained into the overall health status and ecological dynamics of this endangered fish species, and important baseline data on the prevalence, intensity and abundance of parasites infecting the southern pygmy perch will be provided.
Materials and methods
Study site and sample collection
The study was conducted at a farm dam located at 35.862745°S, 147.252098°E. This particular population of southern pygmy perch is a translocated one, originating from fish rescued from a drying lake in the catchment and relocated to the farm dam in September 2014. The fish population has since established a self-sustaining population. On 5 February 2019, a mortality event occurred during a routine monitoring event using bait traps. Bait traps consist of a small collapsible mesh ‘box’ with external dimensions of 245 × 245 × 400 mm, with a single funnel entrance at each end. The mesh size used is ~2.5 mm and the funnel entrances are ~35 mm in diameter. Traps were unbaited and left overnight (set late afternoon, retrieved the following morning). The bait traps were set overnight and retrieved the following morning, with a majority of the trapped fish found dead. The cause of death was likely attributed to low water oxygen concentration resulting from diurnal turnover or water movement within the dam, preventing the fish from accessing suitable oxygen concentrations. No other mortalities were observed elsewhere within the dam.
Fish examination
In total, 81 dead fish were examined for parasite infections. All fish were examined on 6 and 7 February 2019. Postmortem examination was initially performed, followed by the incubation method as described by Shamsi and Suthar (2016) to enhance parasite recovery. The study had approval of the Charles Sturt University’s Animal Ethics Committee (Approval number A19024).
Morphological examination
Parasites collected from the fish were subjected to morphological examination by using light microscopy to capture their characteristic features, as previously described (Shamsi et al. 2019). Identification of the nematode larvae was based on the taxonomic descriptions of nematode larvae provided by Moravec (1998), Moravec (1994) and (Shamsi et al. 2018a). All parasite specimens found in this study were deposited in the South Australian Natural History Museum.
Molecular examination
Genomic DNA (gDNA) was extracted from the collected parasites by using a modified protocol based on the manufacturer’s instructions (Qiagen) (Shamsi et al. 2018b). Polymerase chain-reaction (PCR) amplification of the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) was performed using the primer sets NC16 and NC2, following the conditions described previously (Shamsi et al. 2021b). The PCR products were sent to the Australian Genome Research Facility (AGRF) for sequencing by using the same primers as for the PCR. Sequence chromatograms were quality checked using SeqMan (ver. 8.1.0, DNASTAR, Inc.), and primer sequences were removed for downstream analysis.
Sequence analysis
The obtained ITS-1 sequences from the nematode larvae (GenBank accession: OR186188) were compared with known sequences available in GenBank for identification purposes (Table 1). Pairwise genetic distances between the sequences were calculated using MEGA (ver. 11.0.10, see https://www.megasoftware.net; Kumar et al. 2018), excluding gaps. Phylogenetic analysis was performed using MrBayes (ver. 3.2, see http://nbisweden.github.io/MrBayes/index.html; Ronquist and Huelsenbeck 2003) with the HKY + G model, as determined by jModelTest2 (ver. 2.0, see https://github.com/ddarriba/jmodeltest2; Darriba et al. 2012). The analysis was run for 2 000 000 generations until the standard deviation of split frequencies reached below 0.05. Habronema muscae (KX868082; Jian et al. 2017), another member of suborder Spirurina other than Habronematidae family, was used as an outgroup.
| Taxon (developmental stage) | GenBank accession number | Host (common name followed by scientific name) | Locality | Reference |
|---|---|---|---|---|
| Spiroxys sp. (larva) | OR186188 | Southern pygmy perch, Nannoperca australis | Australia | Present study |
| Spiroxys ankarafantsika (adult) | MW550279 | Pelusios castanoides P. sinuatus P. subniger | Mozambique, South Africa | Nel et al. (2021) |
| Spiroxys sp. (larva) | MH843727 | Killifish, family: Nothobranchiidae | Mozambique | Unpublished |
| Spiroxys japonica | KF530321 | Pelophylax nigromaculatus | China | Li et al. (2014) |
| Spiroxys japonica | KF530322 | Pelophylax nigromaculatus | China | Li et al. (2014) |
| Spiroxys japonica | KF530323 | Pelophylax nigromaculatus | China | Li et al. (2014) |
| Spiroxys japonica | KF530324 | Pelophylax nigromaculatus | Japan | Li et al. (2014) |
| Spiroxys japonica | KF530325 | Lithobates catesbeianus | Japan | Li et al. (2014) |
| Spiroxys hanzaki | KF530326 | Andrias japonicus | Japan | Li et al. (2014) |
| Spiroxys hanzaki, (adults and larva) | LC605542 | Hybrid Andrias between A. japonicus and Chinese Andrias sp. | Japan | Tsuchida et al. (2021) |
| Gnathostoma nipponicum (adult) | AB181157 | Weasel, Mustela sibirica | Japan | Ando et al. (2006) |
| Gnathostoma spinigerum | AB181155 | Swamp eel, Fluta alba | Thailand | Ando et al. (2006) |
| Gnathostoma binucleatum (larva) | AB181159 | Fish, Rhamdia cinerascens | Ecuador | Ando et al. (2006) |
| Gnathostoma doloresi (adult) | AB181156 | Wild boar, Sus scrofa | Japan | Ando et al. (2006) |
| Gnathostoma hispidum (adult) | AB181158 | Pig stomach from larvae in loaches imported from China | Japan | Ando et al. (2006) |
| Echinocephalus sp. (larva) | MW136167 | Rhabdosargus sarba | Australia | Shamsi et al. (2021b) |
| Echinocephalus sp. (larva) | MW136169 | Acanthopagrus australis | Australia | Shamsi et al. (2021b) |
| Echinocephalus sp. (larva) | MW136168 | Rhabdosargus sarba | Australia | Shamsi et al. (2021b) |
| Habronema muscae | KX868082 | Donkey | China | Jian et al. (2017) |
Data analysis
The prevalence of parasite infection was calculated as the percentage of infected fish of the total examined. Mean intensity represents the average number of parasites per infected fish, and mean abundance represents the average number of parasites per examined fish. These parameters were calculated following the methods described by Bush et al. (1997).
Results
Post-mortem examination alone did not show any parasites. However, employing the incubation method resulted in the recovery of nematode larvae in 14 fish, indicating an overall prevalence of 17.3%. The number of nematode larvae per infected fish ranged from one to four, with the majority of fish (n = 9) harbouring only one larva. One fish was found to be infected with four larvae. The mean intensity, representing the average number of parasites per infected fish, was 1.6, whereas the mean abundance, representing the average number of parasites per examined fish, was 0.28.
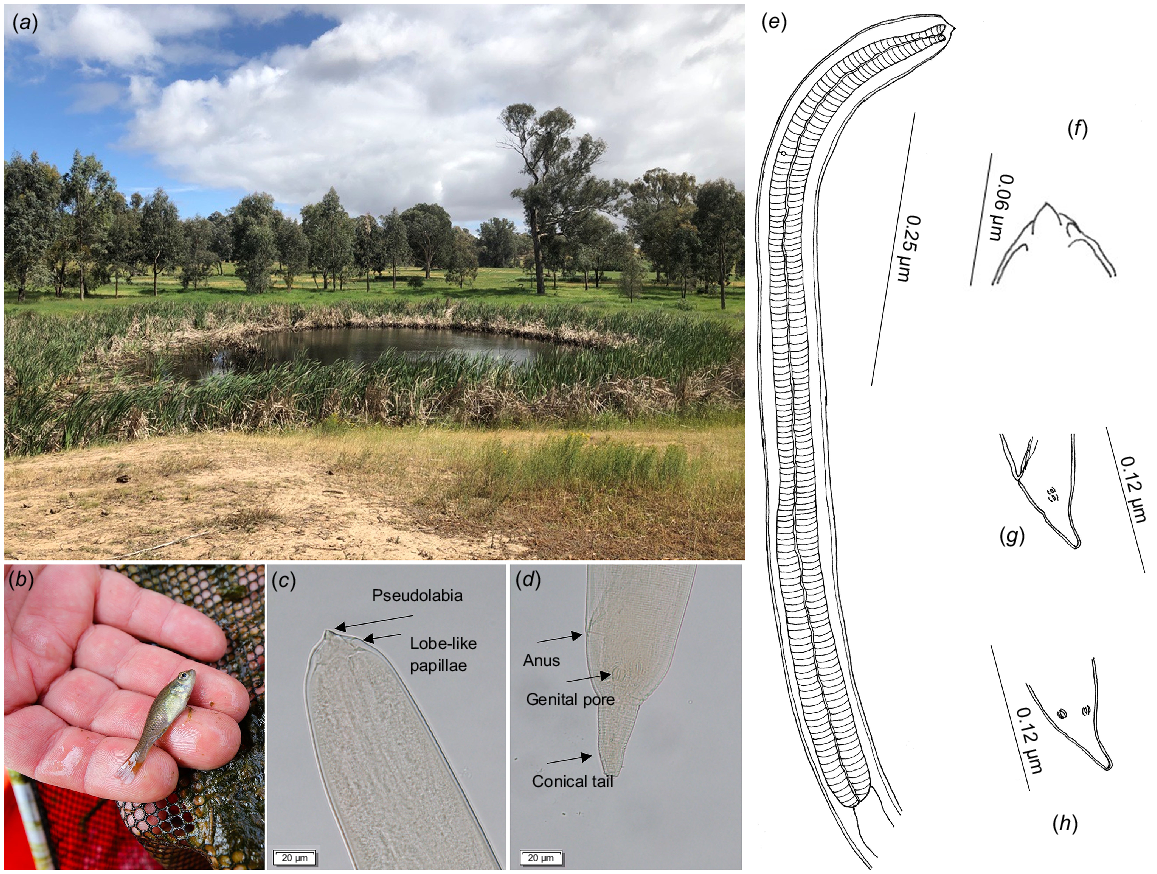
Morphological examination of the nematode larvae confirmed their identification as Spiroxys larvae. The larvae exhibited a finely striated cuticle and distinct triangular lateral pseudolabia (Fig. 1c, f). The oesophagus was composed of muscular (Fig. 1e) and glandular regions. Biometric measurements of the larvae were as follows (average followed by range and number examined): total body length 1.61 mm (0.77–2.18 mm; n = 19), maximum body width 1.61 mm (0.77–2.18 mm; n = 19), nerve ring to the anterior end 0.12 mm (0.09–0.15 mm; n = 3), deirids 0.20 mm (0.16–0.25 mm; n = 5), excretory pore 0.15 mm (0.15–0.15 mm; n = 2) from the anterior end, tail 0.07 mm (0.04–0.13 mm; n = 19), and genital pore (Fig. 1d) 0.05 mm (0.04–0.06 mm; n = 10) from the tip of the tail (Fig. 1d, g, h).
Showing (a) the habitat, (b) the host and (c–h) the parasite, Spiroxys sp. larva in the present study. (c) and (d) are light-microscopy images of the anterior and posterior ends of the larval nematode respectively; (e–h) are illustrations of taxonomically important features of the larval nematode, including, anterior section and eosophagus (e), mouth part (f), lateral view of the posterior end (g), and ventral view of the tails region (h). Terminology follows Truong and Bullard (2021). Note, image (f) corresponds with image (c), and image (g) with image (d).
Molecular analysis targeting the ITS-1 region of the rDNA successfully obtained sequences from two nematode larva specimens (Specimen numbers: 50-1-1, 49-1-1). However, no identical sequences were found in GenBank for comparison (Table 2). Phylogenetic analysis (Fig. 2) confirmed the assignment of the specimens to the family Gnathostomatidae and the genus Spiroxys.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 OR186188A | 47 | 47 | 44 | 44 | 43 | 43 | 44 | 69 | 69 | 150 | 145 | 146 | 145 | 148 | 143 | 143 | 143 | 141 | |
| 2 MW550279 | 14.87 | 0 | 47 | 46 | 45 | 45 | 46 | 65 | 65 | 142 | 144 | 142 | 140 | 144 | 146 | 146 | 146 | 148 | |
| 3 MH843727 | 14.87 | 0.00 | 47 | 46 | 45 | 45 | 46 | 65 | 65 | 142 | 144 | 142 | 140 | 144 | 146 | 146 | 146 | 148 | |
| 4 KF530321 | 13.92 | 14.87 | 14.87 | 2 | 2 | 2 | 1 | 74 | 74 | 143 | 140 | 140 | 143 | 143 | 144 | 144 | 144 | 146 | |
| 5 KF530322 | 13.92 | 14.56 | 14.56 | 0.63 | 2 | 2 | 1 | 75 | 75 | 142 | 139 | 139 | 144 | 142 | 142 | 142 | 142 | 147 | |
| 6 KF530324 | 13.61 | 14.24 | 14.24 | 0.63 | 0.63 | 0 | 1 | 73 | 73 | 144 | 141 | 141 | 144 | 144 | 144 | 144 | 144 | 145 | |
| 7 KF530325 | 13.61 | 14.24 | 14.24 | 0.63 | 0.63 | 0.00 | 1 | 73 | 73 | 144 | 141 | 141 | 144 | 144 | 144 | 144 | 144 | 145 | |
| 8 KF530323 | 13.92 | 14.56 | 14.56 | 0.32 | 0.32 | 0.32 | 0.32 | 74 | 74 | 143 | 140 | 140 | 143 | 143 | 143 | 143 | 143 | 146 | |
| 9 LC605542 | 21.84 | 20.57 | 20.57 | 23.42 | 23.73 | 23.10 | 23.10 | 23.42 | 0 | 159 | 156 | 158 | 153 | 159 | 153 | 153 | 153 | 142 | |
| 10 KF530326 | 21.84 | 20.57 | 20.57 | 23.42 | 23.73 | 23.10 | 23.10 | 23.42 | 0.00 | 159 | 156 | 158 | 153 | 159 | 153 | 153 | 153 | 142 | |
| 11 AB181157 | 47.47 | 44.94 | 44.94 | 45.25 | 44.94 | 45.57 | 45.57 | 45.25 | 50.32 | 50.32 | 21 | 12 | 27 | 17 | 173 | 173 | 173 | 151 | |
| 12 AB181155 | 45.89 | 45.57 | 45.57 | 44.30 | 43.99 | 44.62 | 44.62 | 44.30 | 49.37 | 49.37 | 6.65 | 17 | 31 | 22 | 171 | 171 | 171 | 149 | |
| 13 AB181159 | 46.20 | 44.94 | 44.94 | 44.30 | 43.99 | 44.62 | 44.62 | 44.30 | 50.00 | 50.00 | 3.80 | 5.38 | 33 | 19 | 169 | 169 | 169 | 152 | |
| 14 AB181156 | 45.89 | 44.30 | 44.30 | 45.25 | 45.57 | 45.57 | 45.57 | 45.25 | 48.42 | 48.42 | 8.54 | 9.81 | 10.44 | 28 | 173 | 173 | 173 | 151 | |
| 15 AB181158 | 46.84 | 45.57 | 45.57 | 45.25 | 44.94 | 45.57 | 45.57 | 45.25 | 50.32 | 50.32 | 5.38 | 6.96 | 6.01 | 8.86 | 174 | 174 | 174 | 154 | |
| 16 MW136167 | 45.25 | 46.20 | 46.20 | 45.57 | 44.94 | 45.57 | 45.57 | 45.25 | 48.42 | 48.42 | 54.75 | 54.11 | 53.48 | 54.75 | 55.06 | 0 | 0 | 175 | |
| 17 MW136169 | 45.25 | 46.20 | 46.20 | 45.57 | 44.94 | 45.57 | 45.57 | 45.25 | 48.42 | 48.42 | 54.75 | 54.11 | 53.48 | 54.75 | 55.06 | 0.00 | 0 | 175 | |
| 18 MW136168 | 45.25 | 46.20 | 46.20 | 45.57 | 44.94 | 45.57 | 45.57 | 45.25 | 48.42 | 48.42 | 54.75 | 54.11 | 53.48 | 54.75 | 55.06 | 0.00 | 0.00 | 175 | |
| 19 KX868082 | 44.62 | 46.84 | 46.84 | 46.20 | 46.52 | 45.89 | 45.89 | 46.20 | 44.94 | 44.94 | 47.78 | 47.15 | 48.10 | 47.78 | 48.73 | 55.38 | 55.38 | 55.38 |
ASpecimen obtained in the present study.
Phylogenetic tree based on ITS sequences, showing the placement of our specimen with other Spiroxys spp. Habronema muscae was used as an outgroup. Bayesian posterior probability values of >90% are indicated above the branches (Ando et al. 2006; Li et al. 2014; Nel et al. 2021; Shamsi et al. 2021c; Tsuchida et al. 2021). The asterisk (*) indicates the specimen obtained from this study.
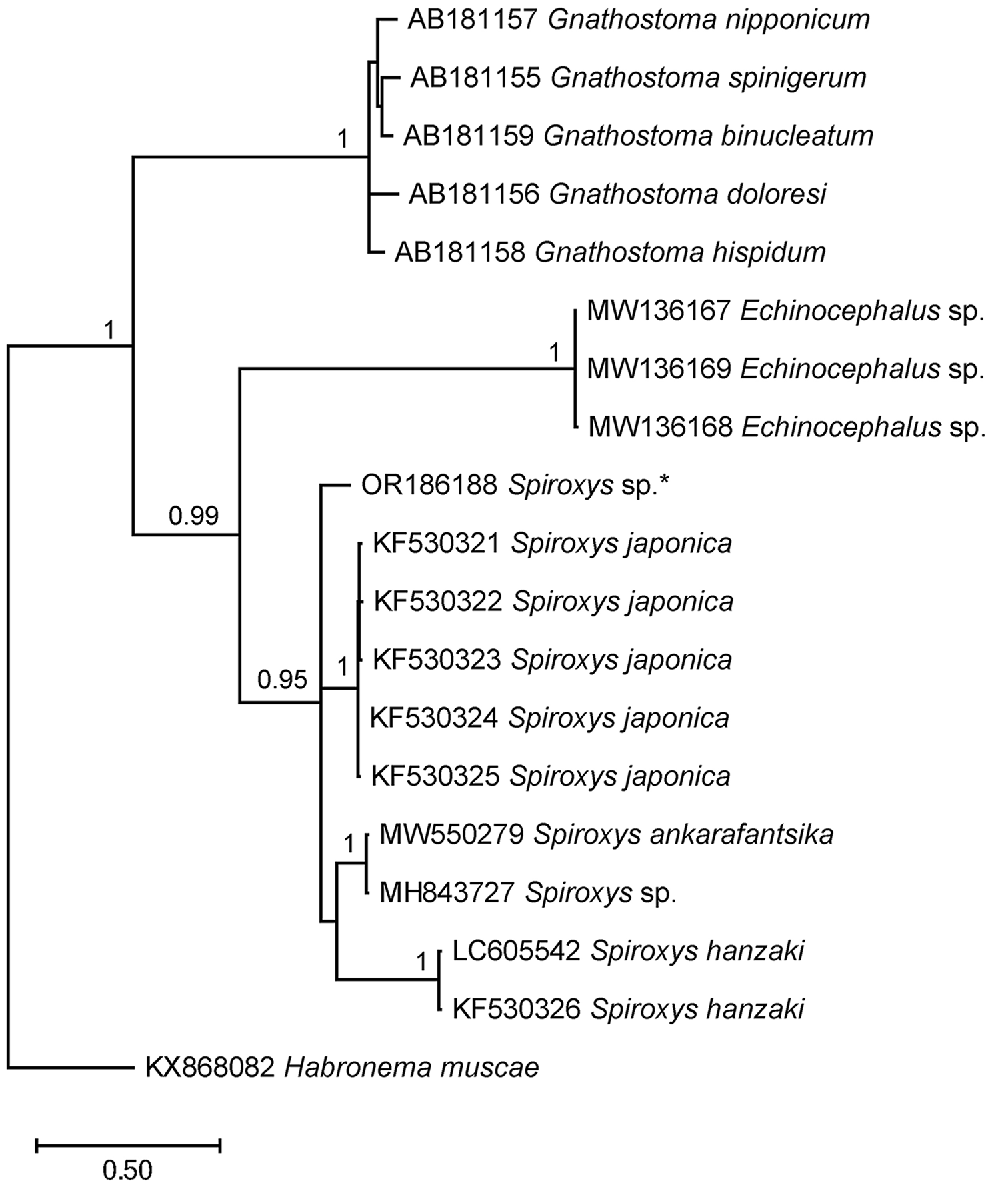
The phylogenetic analysis showed that the nematode larvae found in this study represent a unique genetic lineage within the Spiroxys genus. Owing to the absence of identical sequences from well-identified adult specimens, the specific identity of these larvae remains unknown at this stage.
Discussion
The present study has provided important insights into the occurrence and identification of Spiroxys nematode larvae in southern pygmy perch (N. australis) from a farm dam in south-eastern Australia. The mortality event that occurred in February 2019 prompted the investigation of potential parasitic infections in the affected fish population. During the comprehensive examination of the fish, it was determined that only nematode larvae were present in the examined specimens. Through a combination of morphological and molecular techniques, the nematode larvae were identified as belonging to the genus Spiroxys, representing a unique genetic lineage within the genus.
The values for prevalence, mean intensity and mean abundance of Spiroxys larvae indicated a low infection rate and parasite load in the studied population. However, considering the potential negative effects of parasitic infections on host fitness and population dynamics, further investigations are warranted to assess the potential consequences of Spiroxys infection on the southern pygmy perch population. Furthermore, it is well-documented that larval parasites have the ability to alter the behaviour of their intermediate fish hosts so as to facilitate their transmission to definitive hosts and complete their life cycle as adults (Shamsi et al. 2021c; Freire et al. 2022). Consequently, an important avenue for future research would involve investigating the effect of Spiroxys larval infection on fish behaviour. Understanding how these parasites influence the behaviour of their hosts will provide valuable insights into the ecological dynamics of the parasite–host relationship and contribute to our overall understanding of the complex interactions between parasites and their hosts.
The morphological characteristics observed in the nematode larvae, including the finely striated cuticle, triangular lateral pseudolabia, and the presence of muscular and glandular regions in the oesophagus, align with the description of Spiroxys larvae provided by previous studies (Moravec 1994; Moravec 1998). The biometric measurements of the larvae also corresponded to the reported size ranges for Spiroxys larvae. However, the precise species identification of the larvae could not be established because of the lack of identical sequences in GenBank for comparison. Further investigations involving the identification of adult Spiroxys specimens from the same location are required to confirm the species identity of the larvae.
The phylogenetic analysis showed that the Spiroxys larva in this study clustered within the family Gnathostomatidae and formed a distinct genetic lineage within the Spiroxys genus.
This finding may suggest the existence of potential cryptic diversity within Spiroxys species. It also could be a geographical variation for the parasite, given that there are no other sequences from an Australian representative in the GenBank. Nevertheless, the findings highlighted the need for comprehensive taxonomic and genetic studies to elucidate the species boundaries and evolutionary relationships within the genus. Additional sampling efforts and the inclusion of adult Spiroxys specimens from different geographic locations will contribute to a better understanding of the diversity and distribution of Spiroxys species in Australia.
Nematodes belonging to the genus Spiroxys Schneider are mainly parasites of freshwater chelonians, frogs, salamander, and snakes (Berry 1985) in adult stage, some of which are also commonly found in the locality where these fish were collected in the present study. Of the 18 species assigned to the genus Spiroxys (Baker 1987; Hasegawa et al. 1998; Moravec and Vargas-Vazquez 1998; Roca et al. 2007; Nel et al. 2021), one has been reported in Australia (Berry 1985). The Australian Spiroxys species, S. chelodinae, in its adult form, occurs in three species of freshwater chelonian turtles, namely Chelodina longicollis, C. expansa and C. oblonga, and has been reported from Armidale (New South Wales), Fraser Island (Queensland), Milang, Tailem Bend, Mannum and Lake Alexandrina (South Australia). Australian freshwater turtles eat insects, tadpoles, small freshwater fish, fresh and saltwater prawns and yabbies, snails and mussels and worms. Chelodina longicollis has been sighted in the area of the fish collection in the present study. The presence of Spiroxys larva in the farm dam, where southern pygmy perch was collected, raises the possibility of an association between the fish and freshwater chelonians. Therefore, it is plausible that the Spiroxys larvae found in the southern pygmy perch may represent the larval stage of S. chelodinae, suggesting a potential transmission pathway involving chelonians as intermediate hosts. Further investigations, including the examination of chelonians in the vicinity and the molecular confirmation of the adult Spiroxys species, are required to confirm this hypothesis.
Globally, Spiroxys spp. larvae have been reported in various freshwater copepods, fishes and tadpoles (Moravec 1994). The present study is the first report of the larval stage of the genus in Australia, which shows the role of a native fish, southern pygmy perch, in its life cycle. According to Santos-Clapp et al. (2022), Spiroxys larvae have a higher incidence in smaller fish. Fish may acquire the infection by eating infected first intermediate hosts or paratenic hosts, such as copepods, aquatic insects and molluscs (Moravec 1998).
The detection of Spiroxys larvae in southern pygmy perch raises important ecological and conservation implications. As a native fish species, the southern pygmy perch plays a significant role in maintaining ecosystem balance and functioning in freshwater habitats. Parasitic infections, even at low prevalence and intensity, can influence the health and fitness of host individuals, potentially affecting their reproductive success, growth, and survival.
Understanding the factors influencing the occurrence and transmission of Spiroxys larvae in southern pygmy perch populations is crucial for effective conservation and management strategies. Factors such as habitat characteristics, water quality, host population dynamics, and the presence of intermediate hosts need to be further investigated to elucidate the epidemiology and transmission dynamics of Spiroxys infections in this system. Additionally, assessing the potential effects of Spiroxys infection on the fitness and population dynamics of southern pygmy perch, as well as its interaction with other stressors such as habitat degradation and climate change, is essential for guiding conservation efforts and maintaining the long-term viability of this native fish species.
Nematode larvae were the only parasite found in the present study, which could be due to way we examined the fish. External parasites, including Protozoa and Monogenea, usually leave the host after its death or after change in their environmental conditions and it is possible that we missed the opportunity to find more parasites in them.
Despite facing significant long-term threats, fish species that lack commercial or recreational value often are overlooked in conservation management efforts (Todd et al. 2017). Their exclusion from conservation focus poses a risk to their survival and undermines efforts to safeguard their future. Recognising the importance of these species and addressing the challenges they face is crucial for maintaining biodiversity and promoting effective conservation strategies. Effective management of threatened species requires a thorough understanding of their ecology, as well as the underlying factors that pose threats to their well-being and contribute to disease susceptibility.
References
Ando K, Tsunemori M, Akahane H, Tesana S, Hasegawa H, Chinzei Y (2006) Comparative study on DNA sequences of ribosomal DNA and cytochrome c oxidase subunit 1 of mitochondrial DNA among five species of gnathostomes. Journal of Helminthology 80(1), 7-13.
| Crossref | Google Scholar |
Berry GN (1985) A new species of the genus Spiroxys (Nematoda; Spiruroidea) from Australian chelonians of the genus Chelodina (Chelidae). Systematic Parasitology 7(1), 59-68.
| Crossref | Google Scholar |
Bray DJ, Thompson VJ (2022) Nannoperca australis in Fishes of Australia. Available at https://fishesofaustralia.net.au/home/species/1828 [Verified 16 May 2023]
Bush AO, Lafferty KD, Lotz JM, Shostak AW (1997) Parasitology meets ecology on its own terms: Margolis et al. revisited. The Journal of Parasitology 83(4), 575-583.
| Crossref | Google Scholar |
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9(8), 772.
| Crossref | Google Scholar |
Freire R, Rogers L, Creece D, Shamsi S (2022) Neophobic behavioural responses of parasitised fish to a potential predator and baited hook. Applied Animal Behaviour Science 254, 105722.
| Crossref | Google Scholar |
Habibi F, Shamsi S (2018) Preliminary report of occurrence of Corynosoma spp. (Acanthocephala: Polymorphidae) in Southern Caspian sprat (Clupeonella grimmi). Parasitology Research 117(10), 3327-3331.
| Crossref | Google Scholar |
Hasegawa H, Miyata A, Doi T (1998) Spiroxys hanzaki n. sp. (Nematoda: Gnathostomatidae) collected from the giant salamander, Andrias japonicus (Caudata: Cryptobranchidae), in Japan. The Journal of Parasitology 84(4), 831-834.
| Crossref | Google Scholar |
Jian R, Wang S-W, Zhang W-X, Zhang L-P (2017) Morphological and molecular identification of Habronema spp. (Nematoda: Habronematidae) from donkeys in Xinjiang, China, and notes on the taxonomical status of Habronema majus (Creplin, 1849) and H. microstoma (Schneider, 1866). Systematic Parasitology 94(4), 511-525.
| Crossref | Google Scholar |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35(6), 1547-1549.
| Crossref | Google Scholar |
Li L, Hasegawa H, Roca V, Xu Z, Guo Y-N, Sato A, Zhang L-P (2014) Morphology, ultrastructure and molecular characterisation of Spiroxys japonica Morishita, 1926 (Spirurida: Gnathostomatidae) from Pelophylax nigromaculatus (Hallowell) (Amphibia: Ranidae). Parasitology Research 113(3), 893-901.
| Crossref | Google Scholar |
Mascarenhas CS, Silva RZ, Muller G (2021) Helminth’s assemblage of Trachemys dorbigni (Testudines: Emydidae) in southern Brazil: implications of anthropogenic environments and host’s genders. Iheringia. Serie Zoologia 111, e2021011.
| Crossref | Google Scholar |
Moravec F, Vargas-Vazquez J (1998) Some endohelminths from the freshwater turtle Trachemys scripta from Yucatan, Mexico. Journal of Natural History 32(3), 455-468.
| Crossref | Google Scholar |
Morris SA, Pollard DA, Gehrke PC, Pogonoski JJ (2001) Threatened and potentially threatened freshwater fishes of coastal New South Wales and the Murray–Darling Basin: report to Fisheries Action Program and World Wide Fund for Nature. Final report series number 33, Project Number AA 0959.98. (NSW Fisheries: Sydney, NSW, Australia) Available at https://www.dpi.nsw.gov.au/__data/assets/pdf_file/0010/545617/FFRS-33_Morris-et-al-2001.pdf
Nel T, du Preez L, Netherlands E, Syrota Y, Svitin R (2021) Spiroxys ankarafantsika Roca et Garcia, 2008 (Nematoda: Gnathostomatidae) and other nematodes parasitising Pelusios spp. (Testudines: Pelomedusidae) from South Africa and Mozambique. Acta Parasitologica 66(3), 954-961.
| Google Scholar |
Pearce L, Bice C, Whiterod N, Raadik T (2019) Southern pygmy perch Nannoperca australis. In ‘The IUCN Red List of Threatened Species 2019’. e.T123358579A123382811. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/123358579/123382811
Roca V, Garcia G, Montesinos A (2007) Gastrointestinal helminths found in the three freshwater turtles (Erymnochelys madagascariensis, Pelomedusa subrufa and Pelusios castanoides) from Ankarafantsika National Park, Madagascar. Helminthologia 44(4), 177-182.
| Crossref | Google Scholar |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574.
| Crossref | Google Scholar |
Santos-Clapp MD, Duarte R, Albuquerque MC, Brasil-Sato MC (2022) Helminth endoparasites of endemic fish Pygocentrus piraya (Characiformes, Serrasalmidae) from Tres Marias reservoir, Minas Gerais, Brazil. Anais da Academia Brasileira de Ciencias 94(4), e20201425.
| Crossref | Google Scholar |
Shamsi S, Suthar J (2016) A revised method of examining fish for infection with zoonotic nematode larvae. International Journal of Food Microbiology 227, 13-16.
| Crossref | Google Scholar |
Shamsi S, Turner A, Wassens S (2018a) Description and genetic characterization of a new Contracaecum larval type (Nematoda: Anisakidae) from Australia. Journal of Helminthology 92(2), 216-222.
| Crossref | Google Scholar |
Shamsi S, Steller E, Chen Y (2018b) New and known zoonotic nematode larvae within selected fish species from Queensland waters in Australia. International Journal of Food Microbiology 272, 73-82.
| Crossref | Google Scholar |
Shamsi S, Stoddart A, Smales L, Wassens S (2019) Occurrence of Contracaecum bancrofti larvae in fish in the Murray–Darling Basin. Journal of Helminthology 93(5), 574-579.
| Crossref | Google Scholar |
Shamsi S, Day S, Zhu X, McLellan M, Barton DP, Dang M, Nowak BF (2021a) Wild fish as reservoirs of parasites on Australian Murray Cod farms. Aquaculture 539, 736584.
| Crossref | Google Scholar |
Shamsi S, Steller E, Zhu X (2021b) The occurrence and clinical importance of infectious stage of Echinocephalus (Nematoda: Gnathostomidae) larvae in selected Australian edible fish. Parasitology International 83, 102333.
| Crossref | Google Scholar |
Shamsi S, Rogers L, Sales E, Kopf RK, Freire R (2021c) Do parasites influence behavioural traits of wild and hatchery-reared Murray cod, Maccullochella peelii? Parasitology Research 120(2), 515-523.
| Crossref | Google Scholar |
Shamsi S, Francis N, Masiga J, Barton DP, Zhu X, Pearce L, McLellan M (2023) Occurrence and characterisation of Eustrongylides species in Australian native birds and fish. Food and Waterborne Parasitology 30, e00189.
| Crossref | Google Scholar |
Todd CR, Koehn JD, Pearce L, Dodd L, Humphries P, Morrongiello JR (2017) Forgotten fishes: what is the future for small threatened freshwater fish? Population risk assessment for southern pygmy perch, Nannoperca australis. Aquatic Conservation: Marine and Freshwater Ecosystems 27(6), 1290-1300.
| Crossref | Google Scholar |
Truong TN, Bullard SA (2021) Susceptibility of channel catfish (Ictalurus punctatus), blue catfish (Ictalurus furcatus), and their commercially cultured hybrid to metazoan parasite infection in earthen pond aquaculture. Comparative Parasitology 88(1), 93-112.
| Crossref | Google Scholar |
Tsuchida K, Urabe M, Nishikawa K (2021) The first survey for helminths parasitic in hybrid and introduced giant salamanders, genus Andrias (Amphibia: Caudata: Cryptobranchidae) in Kyoto, Japan. Current Herpetology 40(2), 109-119.
| Crossref | Google Scholar |


